|
Symbol Name ID |
Fut1
fucosyltransferase 1 MGI:109375 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
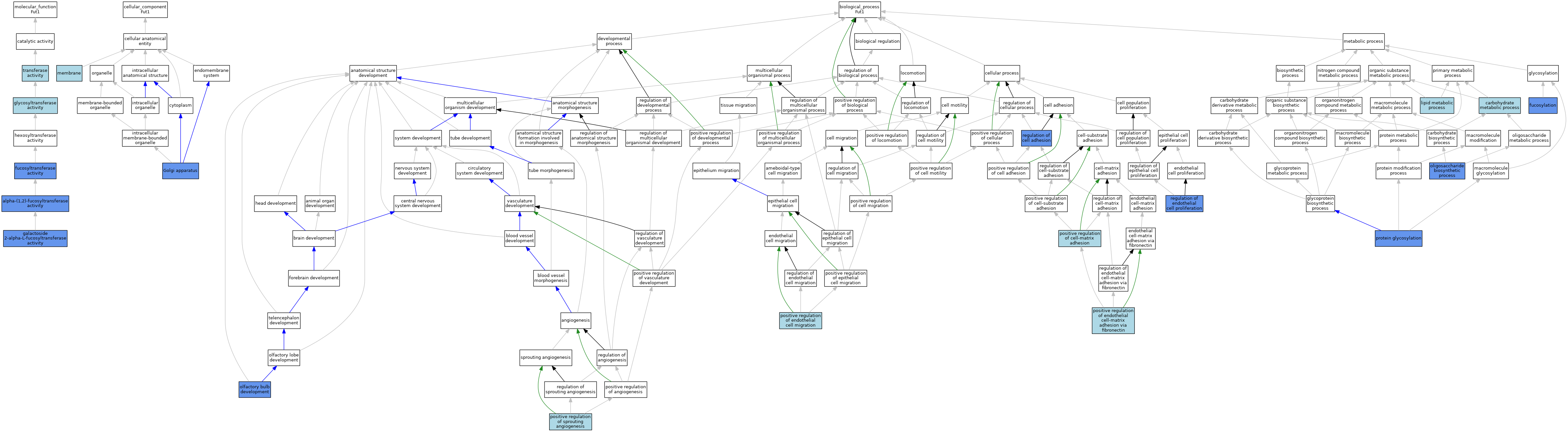
| Molecular Function | GO:0031127 | alpha-(1,2)-fucosyltransferase activity | IDA | J:89929 | |||||||||
| Molecular Function | GO:0031127 | alpha-(1,2)-fucosyltransferase activity | IDA | J:68898 | |||||||||
| Molecular Function | GO:0008417 | fucosyltransferase activity | IMP | J:72946 | |||||||||
| Molecular Function | GO:0008417 | fucosyltransferase activity | IMP | J:72946 | |||||||||
| Molecular Function | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity | IDA | J:43607 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IMP | J:72946 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IMP | J:72946 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0036065 | fucosylation | IDA | J:284978 | |||||||||
| Biological Process | GO:0036065 | fucosylation | IDA | J:89929 | |||||||||
| Biological Process | GO:0036065 | fucosylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0036065 | fucosylation | IDA | J:43607 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0021772 | olfactory bulb development | IMP | J:119285 | |||||||||
| Biological Process | GO:0009312 | oligosaccharide biosynthetic process | IMP | J:72946 | |||||||||
| Biological Process | GO:0009312 | oligosaccharide biosynthetic process | IMP | J:72946 | |||||||||
| Biological Process | GO:0001954 | positive regulation of cell-matrix adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0010595 | positive regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | ISO | J:164563 | |||||||||
| Biological Process | GO:1903672 | positive regulation of sprouting angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006486 | protein glycosylation | IDA | J:68898 | |||||||||
| Biological Process | GO:0006486 | protein glycosylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | IDA | J:284978 | |||||||||
| Biological Process | GO:0001936 | regulation of endothelial cell proliferation | IMP | J:284978 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||