|
Symbol Name ID |
Adm
adrenomedullin MGI:108058 |
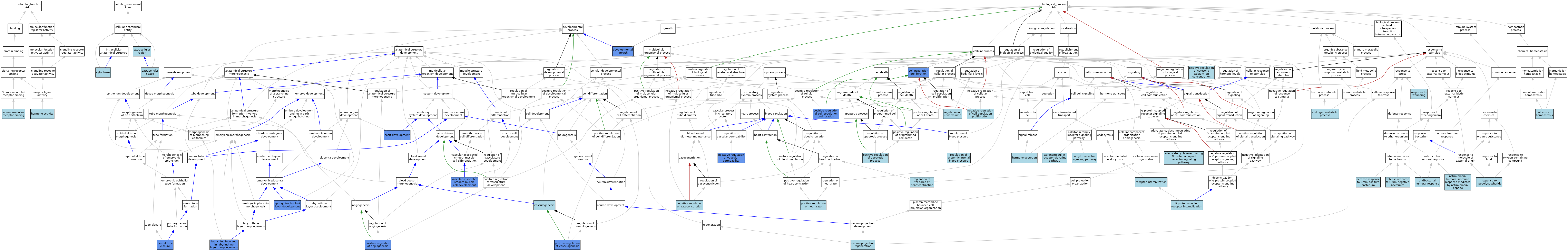
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0031700 | adrenomedullin receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031700 | adrenomedullin receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0031700 | adrenomedullin receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005179 | hormone activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005179 | hormone activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:1990410 | adrenomedullin receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:1990410 | adrenomedullin receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0097647 | amylin receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0008209 | androgen metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | ISO | J:164563 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | ISO | J:164563 | |||||||||
| Biological Process | GO:0060670 | branching involved in labyrinthine layer morphogenesis | IMP | J:114553 | |||||||||
| Biological Process | GO:0055074 | calcium ion homeostasis | ISO | J:155856 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IMP | J:106927 | |||||||||
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0048589 | developmental growth | IMP | J:114553 | |||||||||
| Biological Process | GO:0002031 | G protein-coupled receptor internalization | ISO | J:164563 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:130931 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:106927 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:114553 | |||||||||
| Biological Process | GO:0046879 | hormone secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0043116 | negative regulation of vascular permeability | IMP | J:130931 | |||||||||
| Biological Process | GO:0045906 | negative regulation of vasoconstriction | ISO | J:164563 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IMP | J:114553 | |||||||||
| Biological Process | GO:0031102 | neuron projection regeneration | ISO | J:155856 | |||||||||
| Biological Process | GO:0045766 | positive regulation of angiogenesis | IMP | J:130931 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IMP | J:106927 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0010460 | positive regulation of heart rate | ISO | J:155856 | |||||||||
| Biological Process | GO:0010460 | positive regulation of heart rate | ISO | J:164563 | |||||||||
| Biological Process | GO:0010460 | positive regulation of heart rate | IBA | J:265628 | |||||||||
| Biological Process | GO:2001214 | positive regulation of vasculogenesis | IMP | J:130931 | |||||||||
| Biological Process | GO:0031623 | receptor internalization | ISO | J:164563 | |||||||||
| Biological Process | GO:0003073 | regulation of systemic arterial blood pressure | ISO | J:164563 | |||||||||
| Biological Process | GO:0003073 | regulation of systemic arterial blood pressure | IBA | J:265628 | |||||||||
| Biological Process | GO:0002026 | regulation of the force of heart contraction | ISO | J:155856 | |||||||||
| Biological Process | GO:0035809 | regulation of urine volume | ISO | J:164563 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | ISO | J:155856 | |||||||||
| Biological Process | GO:0009611 | response to wounding | ISO | J:155856 | |||||||||
| Biological Process | GO:0060712 | spongiotrophoblast layer development | IMP | J:114553 | |||||||||
| Biological Process | GO:0097084 | vascular associated smooth muscle cell development | IMP | J:130931 | |||||||||
| Biological Process | GO:0001570 | vasculogenesis | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||