|
Symbol Name ID |
Rhob
ras homolog family member B MGI:107949 |
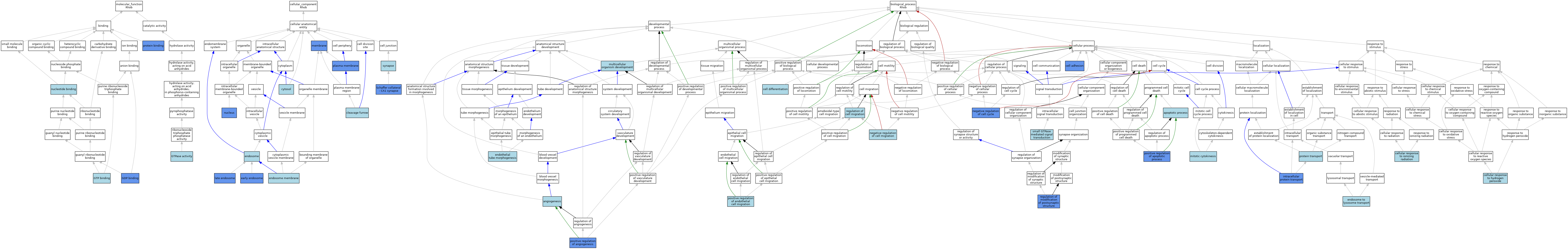
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019003 | GDP binding | IDA | J:36812 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | TAS | J:113699 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:36812 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:30350 | |||||||||
| Cellular Component | GO:0032154 | cleavage furrow | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005769 | early endosome | IDA | J:125001 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0010008 | endosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005770 | late endosome | IDA | J:125001 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:36812 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:86323 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-5625893 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-5666183 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-8981447 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:89351 | |||||||||
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:159018 | |||||||||
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:159018 | |||||||||
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IMP | J:159018 | |||||||||
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:159018 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IDA | J:71577 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0070301 | cellular response to hydrogen peroxide | ISO | J:164563 | |||||||||
| Biological Process | GO:0071479 | cellular response to ionizing radiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008333 | endosome to lysosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0061154 | endothelial tube morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | IMP | J:125001 | |||||||||
| Biological Process | GO:0000281 | mitotic cytokinesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0045786 | negative regulation of cell cycle | IMP | J:71577 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0045766 | positive regulation of angiogenesis | IMP | J:86323 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IDA | J:114053 | |||||||||
| Biological Process | GO:0010595 | positive regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0030334 | regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0099159 | regulation of modification of postsynaptic structure | IDA | J:159018 | |||||||||
| Biological Process | GO:0099159 | regulation of modification of postsynaptic structure | IDA | J:159018 | |||||||||
| Biological Process | GO:0099159 | regulation of modification of postsynaptic structure | IDA | J:159018 | |||||||||
| Biological Process | GO:0099159 | regulation of modification of postsynaptic structure | IMP | J:159018 | |||||||||
| Biological Process | GO:0007264 | small GTPase mediated signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||