|
Symbol Name ID |
Prkcsh
protein kinase C substrate 80K-H MGI:107877 |
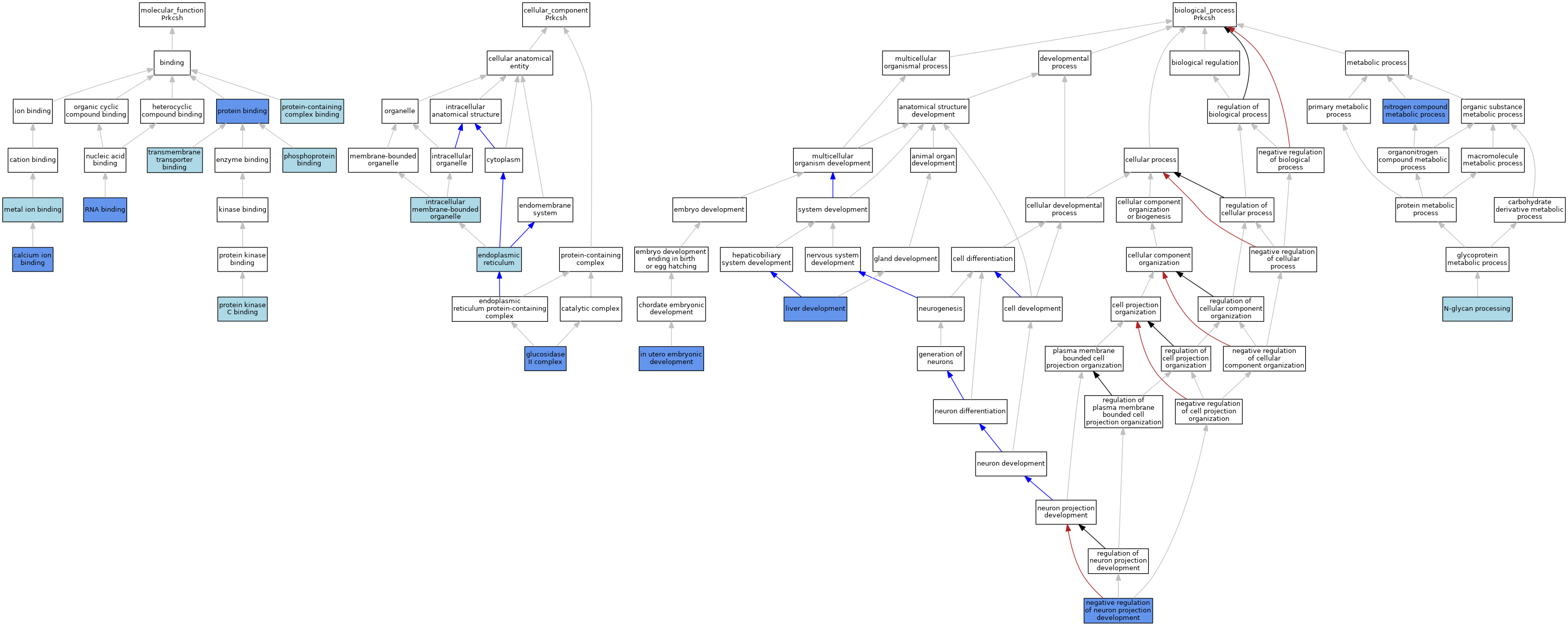
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | IDA | J:261844 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051219 | phosphoprotein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:261844 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:185071 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:40482 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:185071 | |||||||||
| Molecular Function | GO:0005080 | protein kinase C binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IDA | J:108921 | |||||||||
| Molecular Function | GO:0044325 | transmembrane transporter binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | TAS | J:40482 | |||||||||
| Cellular Component | GO:0017177 | glucosidase II complex | IDA | J:261844 | |||||||||
| Cellular Component | GO:0017177 | glucosidase II complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0017177 | glucosidase II complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0017177 | glucosidase II complex | IPI | J:40482 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:188763 | |||||||||
| Biological Process | GO:0001889 | liver development | IBA | J:265628 | |||||||||
| Biological Process | GO:0001889 | liver development | IGI | J:188763 | |||||||||
| Biological Process | GO:0006491 | N-glycan processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0006491 | N-glycan processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0006491 | N-glycan processing | TAS | J:40482 | |||||||||
| Biological Process | GO:0010977 | negative regulation of neuron projection development | IMP | J:185071 | |||||||||
| Biological Process | GO:0006807 | nitrogen compound metabolic process | IGI | J:188763 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||