|
Symbol Name ID |
Kif3b
kinesin family member 3B MGI:107688 |
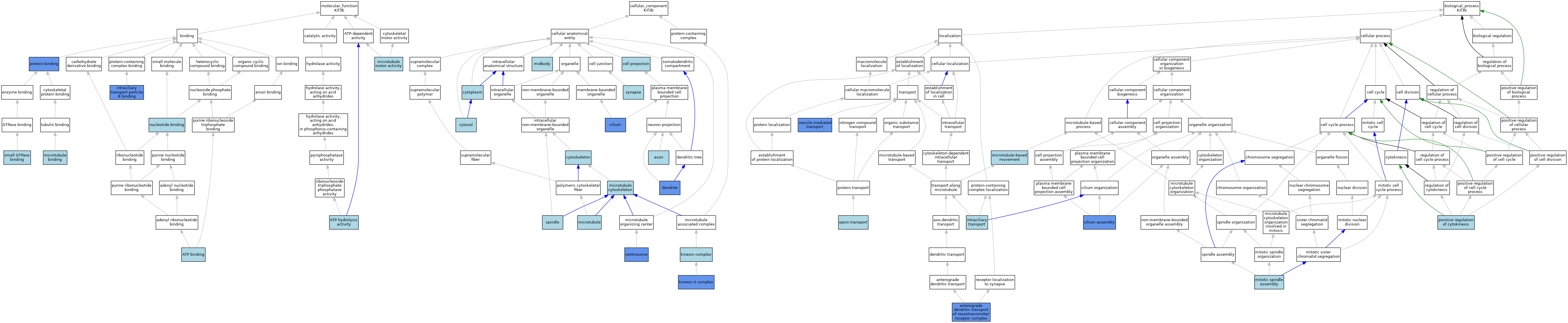
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0120170 | intraciliary transport particle B binding | IDA | J:86875 | |||||||||
| Molecular Function | GO:0120170 | intraciliary transport particle B binding | IDA | J:86875 | |||||||||
| Molecular Function | GO:0120170 | intraciliary transport particle B binding | IDA | J:86875 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003777 | microtubule motor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:248463 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:286227 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:126834 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:207074 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:151268 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:98470 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:98470 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:98470 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:86875 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:98470 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:86875 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030424 | axon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005813 | centrosome | IDA | J:225786 | |||||||||
| Cellular Component | GO:0005813 | centrosome | IDA | J:154656 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:233668 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:51419 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:154656 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-2316350 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-984713 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:286227 | |||||||||
| Cellular Component | GO:0005871 | kinesin complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016939 | kinesin II complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016939 | kinesin II complex | IDA | J:151268 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0015630 | microtubule cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030496 | midbody | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005819 | spindle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEA | J:60000 | |||||||||
| Biological Process | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | IMP | J:286227 | |||||||||
| Biological Process | GO:0060271 | cilium assembly | IMP | J:51419 | |||||||||
| Biological Process | GO:0060271 | cilium assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0042073 | intraciliary transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0007018 | microtubule-based movement | IBA | J:265628 | |||||||||
| Biological Process | GO:0090307 | mitotic spindle assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0036372 | opsin transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0032467 | positive regulation of cytokinesis | ISO | J:155856 | |||||||||
| Biological Process | GO:0016192 | vesicle-mediated transport | IMP | J:286227 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||