|
Symbol Name ID |
Fpr1
formyl peptide receptor 1 MGI:107443 |
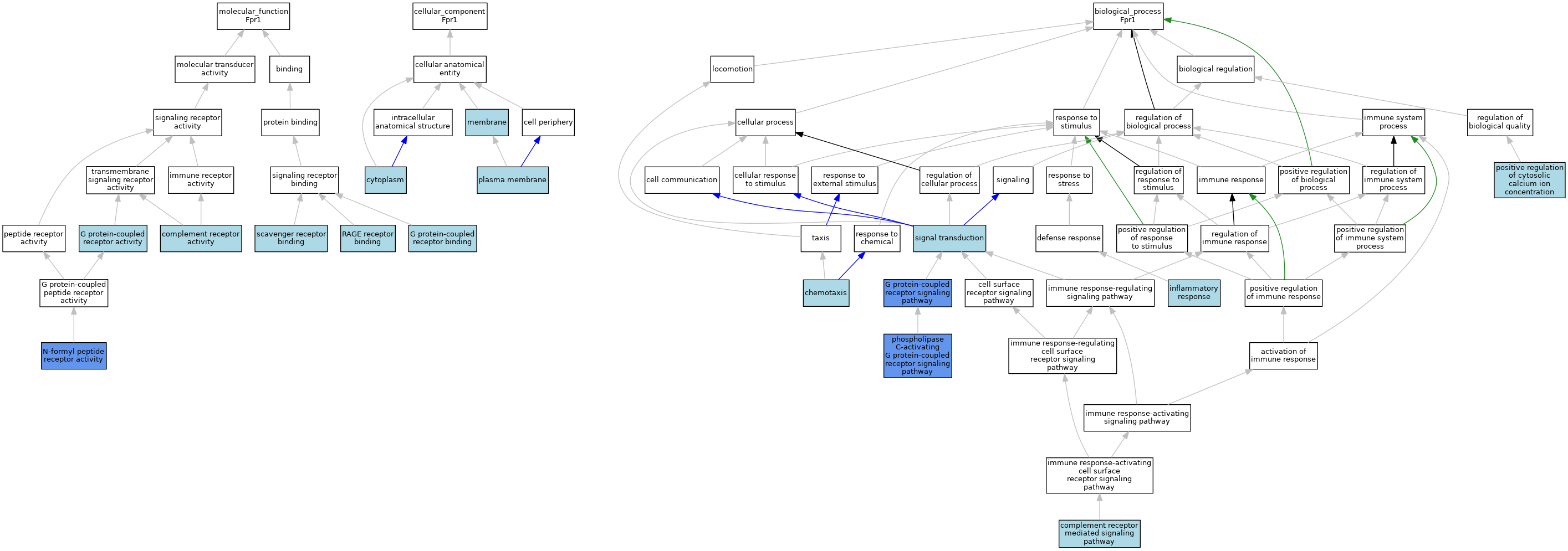
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004875 | complement receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001664 | G protein-coupled receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004982 | N-formyl peptide receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004982 | N-formyl peptide receptor activity | ISO | J:78957 | |||||||||
| Molecular Function | GO:0004982 | N-formyl peptide receptor activity | IDA | J:192967 | |||||||||
| Molecular Function | GO:0004982 | N-formyl peptide receptor activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0050786 | RAGE receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005124 | scavenger receptor binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0006935 | chemotaxis | TAS | J:80798 | |||||||||
| Biological Process | GO:0002430 | complement receptor mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:78957 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IDA | J:80798 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IBA | J:265628 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | ISO | J:73065 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | IDA | J:192967 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | IBA | J:265628 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||