|
Symbol Name ID |
Dnm1
dynamin 1 MGI:107384 |
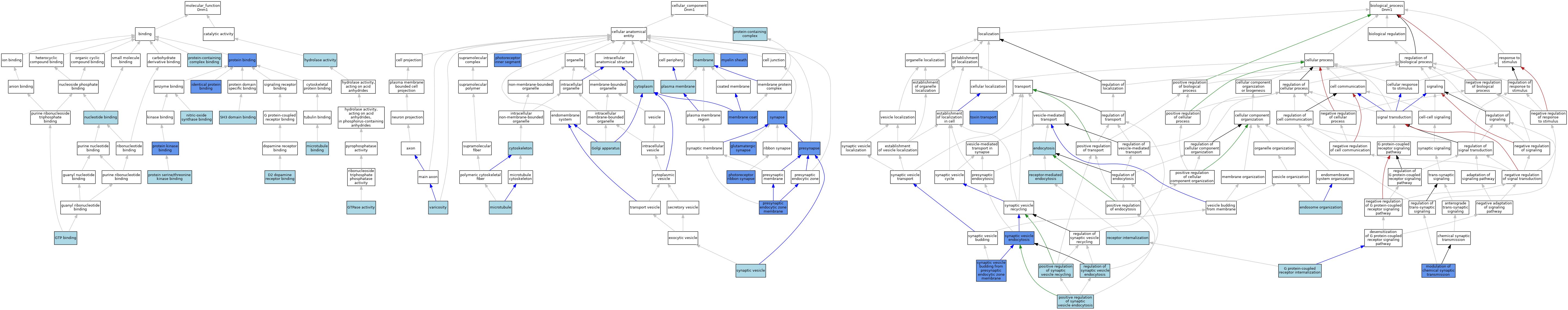
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0031749 | D2 dopamine receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:163311 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0050998 | nitric-oxide synthase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107428 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:209576 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:169453 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:113994 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:204661 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222139 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:152583 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:152583 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:81675 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:214070 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IPI | J:169453 | |||||||||
| Molecular Function | GO:0120283 | protein serine/threonine kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:263390 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:263390 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030117 | membrane coat | IDA | J:75306 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | HDA | J:145263 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:214070 | |||||||||
| Cellular Component | GO:0098684 | photoreceptor ribbon synapse | IDA | J:199646 | |||||||||
| Cellular Component | GO:0098684 | photoreceptor ribbon synapse | IDA | J:199646 | |||||||||
| Cellular Component | GO:0098684 | photoreceptor ribbon synapse | IDA | J:199646 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098793 | presynapse | IDA | J:121111 | |||||||||
| Cellular Component | GO:0098835 | presynaptic endocytic zone membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098835 | presynaptic endocytic zone membrane | IDA | J:199646 | |||||||||
| Cellular Component | GO:0098835 | presynaptic endocytic zone membrane | IDA | J:199646 | |||||||||
| Cellular Component | GO:0098835 | presynaptic endocytic zone membrane | IDA | J:199646 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:121111 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0008021 | synaptic vesicle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043196 | varicosity | ISO | J:155856 | |||||||||
| Biological Process | GO:0006897 | endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0007032 | endosome organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0002031 | G protein-coupled receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | IDA | J:263390 | |||||||||
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | IMP | J:263390 | |||||||||
| Biological Process | GO:1900244 | positive regulation of synaptic vesicle endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:1903423 | positive regulation of synaptic vesicle recycling | ISO | J:155856 | |||||||||
| Biological Process | GO:0031623 | receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:0031623 | receptor internalization | IBA | J:265628 | |||||||||
| Biological Process | GO:0006898 | receptor-mediated endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0006898 | receptor-mediated endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:1900242 | regulation of synaptic vesicle endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | IGI | J:175247 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IDA | J:175247 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IDA | J:175247 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IMP | J:175247 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IDA | J:121111 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IDA | J:121111 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IMP | J:121111 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | IDA | J:121111 | |||||||||
| Biological Process | GO:1901998 | toxin transport | IMP | J:197389 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||