|
Symbol Name ID |
Adam9
ADAM metallopeptidase domain 9 MGI:105376 |
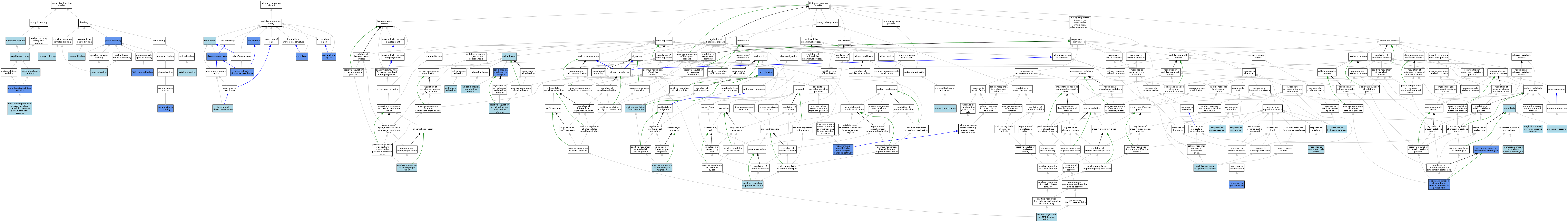
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005518 | collagen binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043236 | laminin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IDA | J:186216 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008237 | metallopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:113785 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:233591 | |||||||||
| Molecular Function | GO:0005080 | protein kinase C binding | IPI | J:51810 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | IDA | J:31451 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:31451 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:97065 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IDA | J:143698 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IMP | J:143698 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:97065 | |||||||||
| Biological Process | GO:0042987 | amyloid precursor protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0033627 | cell adhesion mediated by integrin | IDA | J:233591 | |||||||||
| Biological Process | GO:0033627 | cell adhesion mediated by integrin | ISO | J:164563 | |||||||||
| Biological Process | GO:0016477 | cell migration | IDA | J:233591 | |||||||||
| Biological Process | GO:0033631 | cell-cell adhesion mediated by integrin | IBA | J:265628 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | ISO | J:164563 | |||||||||
| Biological Process | GO:0006509 | membrane protein ectodomain proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006509 | membrane protein ectodomain proteolysis | IDA | J:186216 | |||||||||
| Biological Process | GO:0006509 | membrane protein ectodomain proteolysis | IDA | J:148993 | |||||||||
| Biological Process | GO:0006509 | membrane protein ectodomain proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006509 | membrane protein ectodomain proteolysis | IBA | J:265628 | |||||||||
| Biological Process | GO:0031293 | membrane protein intracellular domain proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0042117 | monocyte activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0033630 | positive regulation of cell adhesion mediated by integrin | ISO | J:164563 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0051549 | positive regulation of keratinocyte migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0034241 | positive regulation of macrophage fusion | ISO | J:164563 | |||||||||
| Biological Process | GO:0043406 | positive regulation of MAP kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | IDA | J:51810 | |||||||||
| Biological Process | GO:0050714 | positive regulation of protein secretion | ISO | J:164563 | |||||||||
| Biological Process | GO:0016485 | protein processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0051592 | response to calcium ion | ISO | J:164563 | |||||||||
| Biological Process | GO:0051384 | response to glucocorticoid | IMP | J:143682 | |||||||||
| Biological Process | GO:0042542 | response to hydrogen peroxide | ISO | J:164563 | |||||||||
| Biological Process | GO:0010042 | response to manganese ion | ISO | J:164563 | |||||||||
| Biological Process | GO:0034612 | response to tumor necrosis factor | ISO | J:164563 | |||||||||
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | IMP | J:143690 | |||||||||
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||