|
Symbol Name ID |
Adra1b
adrenergic receptor, alpha 1b MGI:104774 |
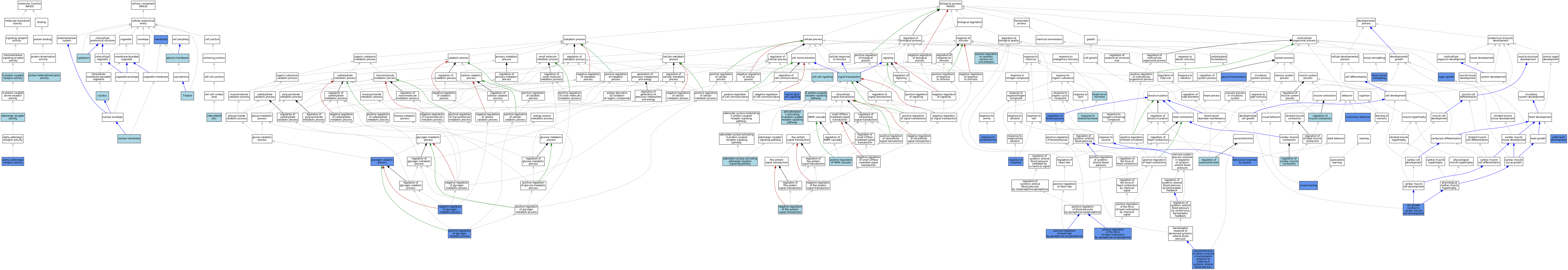
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004935 | adrenergic receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IMP | J:83876 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IMP | J:43591 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IDA | J:76004 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IDA | J:18949 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IMP | J:43591 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IMP | J:43591 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IGI | J:83876 | |||||||||
| Molecular Function | GO:0004937 | alpha1-adrenergic receptor activity | IDA | J:76004 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0014704 | intercalated disc | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:76004 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030315 | T-tubule | ISO | J:155856 | |||||||||
| Biological Process | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007512 | adult heart development | IGI | J:83876 | |||||||||
| Biological Process | GO:0048148 | behavioral response to cocaine | IMP | J:76004 | |||||||||
| Biological Process | GO:0001974 | blood vessel remodeling | IMP | J:95771 | |||||||||
| Biological Process | GO:0061049 | cell growth involved in cardiac muscle cell development | IGI | J:83876 | |||||||||
| Biological Process | GO:0007267 | cell-cell signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0042593 | glucose homeostasis | IMP | J:87718 | |||||||||
| Biological Process | GO:0005980 | glycogen catabolic process | IMP | J:87718 | |||||||||
| Biological Process | GO:0005980 | glycogen catabolic process | IMP | J:87718 | |||||||||
| Biological Process | GO:0007626 | locomotory behavior | IMP | J:68177 | |||||||||
| Biological Process | GO:0045818 | negative regulation of glycogen catabolic process | IMP | J:87718 | |||||||||
| Biological Process | GO:0035024 | negative regulation of Rho protein signal transduction | ISO | J:136004 | |||||||||
| Biological Process | GO:0150099 | neuron-glial cell signaling | IGI | J:273582 | |||||||||
| Biological Process | GO:0035265 | organ growth | IGI | J:83876 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | IBA | J:265628 | |||||||||
| Biological Process | GO:0045819 | positive regulation of glycogen catabolic process | IMP | J:87718 | |||||||||
| Biological Process | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | IBA | J:265628 | |||||||||
| Biological Process | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | IGI | J:83876 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IBA | J:265628 | |||||||||
| Biological Process | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine | IGI | J:83036 | |||||||||
| Biological Process | GO:0008217 | regulation of blood pressure | IMP | J:255410 | |||||||||
| Biological Process | GO:0055117 | regulation of cardiac muscle contraction | IEA | J:72247 | |||||||||
| Biological Process | GO:0006937 | regulation of muscle contraction | IEA | J:72247 | |||||||||
| Biological Process | GO:0019229 | regulation of vasoconstriction | IEA | J:72247 | |||||||||
| Biological Process | GO:0001975 | response to amphetamine | IMP | J:76004 | |||||||||
| Biological Process | GO:0001975 | response to amphetamine | IMP | J:84429 | |||||||||
| Biological Process | GO:0009725 | response to hormone | ISO | J:155856 | |||||||||
| Biological Process | GO:0043278 | response to morphine | IMP | J:76004 | |||||||||
| Biological Process | GO:0048545 | response to steroid hormone | ISO | J:155856 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0001987 | vasoconstriction of artery involved in baroreceptor response to lowering of systemic arterial blood pressure | IMP | J:43591 | |||||||||
| Biological Process | GO:0008542 | visual learning | IMP | J:68177 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||