|
Symbol Name ID |
Kcnj2
potassium inwardly-rectifying channel, subfamily J, member 2 MGI:104744 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
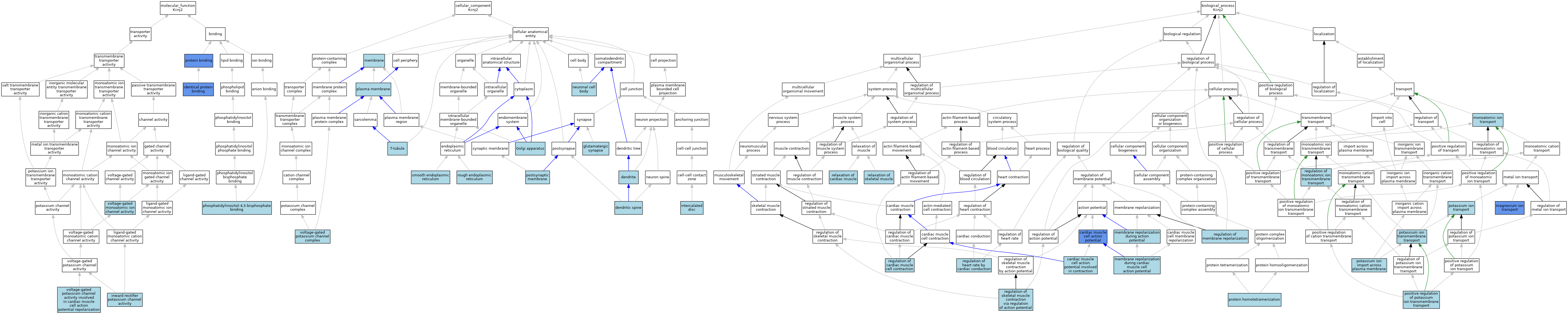
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:177217 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:210915 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005242 | inward rectifier potassium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005242 | inward rectifier potassium channel activity | TAS | Reactome:R-MMU-9729881 | |||||||||
| Molecular Function | GO:0005242 | inward rectifier potassium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:113938 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114059 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:177217 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114113 | |||||||||
| Molecular Function | GO:0005244 | voltage-gated monoatomic ion channel activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043197 | dendritic spine | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0014704 | intercalated disc | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9729881 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005791 | rough endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030315 | T-tubule | ISO | J:155856 | |||||||||
| Cellular Component | GO:0008076 | voltage-gated potassium channel complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0086001 | cardiac muscle cell action potential | IMP | J:94675 | |||||||||
| Biological Process | GO:0086002 | cardiac muscle cell action potential involved in contraction | ISO | J:164563 | |||||||||
| Biological Process | GO:0015693 | magnesium ion transport | IDA | J:18355 | |||||||||
| Biological Process | GO:0086011 | membrane repolarization during action potential | ISO | J:164563 | |||||||||
| Biological Process | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | ISO | J:164563 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:1901381 | positive regulation of potassium ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:1990573 | potassium ion import across plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:1990573 | potassium ion import across plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006813 | potassium ion transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0051289 | protein homotetramerization | ISO | J:164563 | |||||||||
| Biological Process | GO:0086004 | regulation of cardiac muscle cell contraction | ISO | J:155856 | |||||||||
| Biological Process | GO:0086091 | regulation of heart rate by cardiac conduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0060306 | regulation of membrane repolarization | ISO | J:164563 | |||||||||
| Biological Process | GO:0034765 | regulation of monoatomic ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | ISO | J:164563 | |||||||||
| Biological Process | GO:0055119 | relaxation of cardiac muscle | ISO | J:164563 | |||||||||
| Biological Process | GO:0090076 | relaxation of skeletal muscle | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||