|
Symbol Name ID |
Akap1
A kinase anchor protein 1 MGI:104729 |
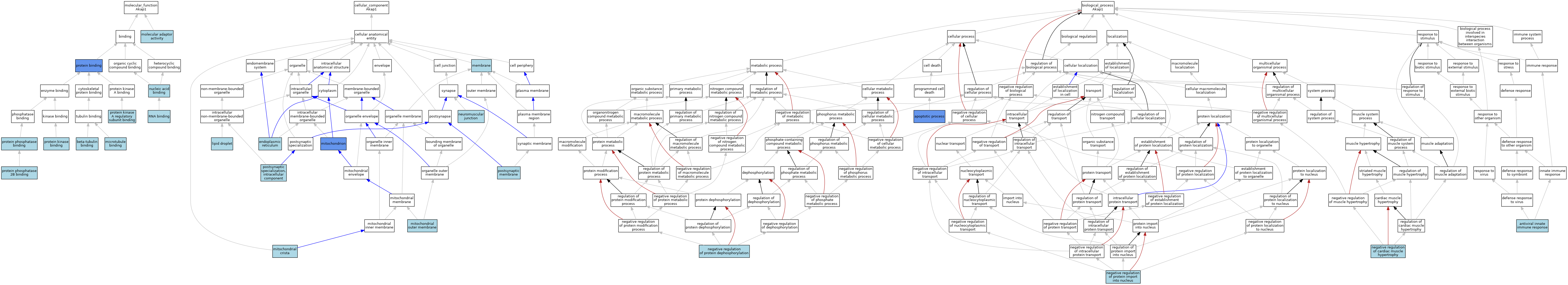
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0048487 | beta-tubulin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200402 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:90992 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:289389 | |||||||||
| Molecular Function | GO:0034237 | protein kinase A regulatory subunit binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0034237 | protein kinase A regulatory subunit binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0030346 | protein phosphatase 2B binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019903 | protein phosphatase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005811 | lipid droplet | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030061 | mitochondrial crista | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005741 | mitochondrial outer membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005741 | mitochondrial outer membrane | TAS | Reactome:R-NUL-8951809 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:289389 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0031594 | neuromuscular junction | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031594 | neuromuscular junction | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0099091 | postsynaptic specialization, intracellular component | ISO | J:155856 | |||||||||
| Biological Process | GO:0140374 | antiviral innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:289389 | |||||||||
| Biological Process | GO:0010614 | negative regulation of cardiac muscle hypertrophy | ISO | J:155856 | |||||||||
| Biological Process | GO:0035308 | negative regulation of protein dephosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0042308 | negative regulation of protein import into nucleus | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||