|
Symbol Name ID |
Ptx3
pentraxin related gene MGI:104641 |
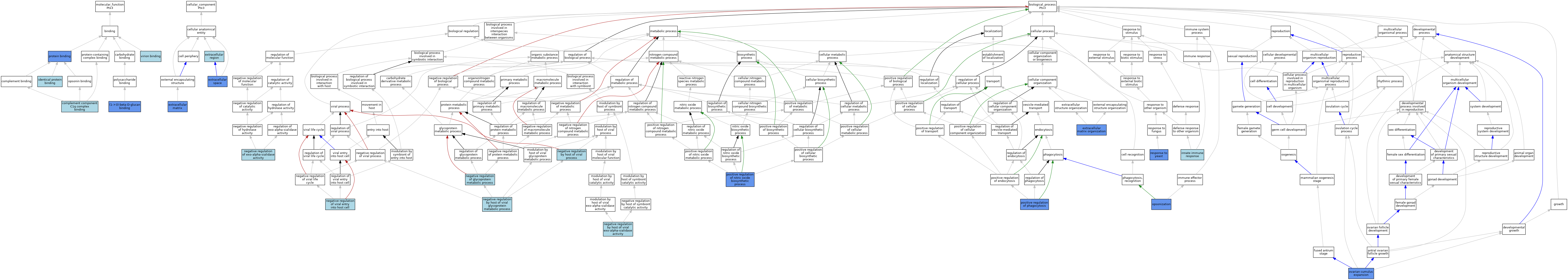
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0001872 | (1->3)-beta-D-glucan binding | IMP | J:89006 | |||||||||
| Molecular Function | GO:0001849 | complement component C1q complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001849 | complement component C1q complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:126717 | |||||||||
| Molecular Function | GO:0046790 | virion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031012 | extracellular matrix | IDA | J:126717 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Biological Process | GO:0030198 | extracellular matrix organization | IMP | J:126717 | |||||||||
| Biological Process | GO:0030198 | extracellular matrix organization | IDA | J:126717 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0044869 | negative regulation by host of viral exo-alpha-sialidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0044871 | negative regulation by host of viral glycoprotein metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0044793 | negative regulation by host of viral process | IBA | J:265628 | |||||||||
| Biological Process | GO:1903016 | negative regulation of exo-alpha-sialidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:1903019 | negative regulation of glycoprotein metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0046597 | negative regulation of viral entry into host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0008228 | opsonization | IMP | J:89006 | |||||||||
| Biological Process | GO:0001550 | ovarian cumulus expansion | IMP | J:126717 | |||||||||
| Biological Process | GO:0001550 | ovarian cumulus expansion | IDA | J:126717 | |||||||||
| Biological Process | GO:0045429 | positive regulation of nitric oxide biosynthetic process | IMP | J:89006 | |||||||||
| Biological Process | GO:0050766 | positive regulation of phagocytosis | IMP | J:89006 | |||||||||
| Biological Process | GO:0001878 | response to yeast | IMP | J:89006 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||