|
Symbol Name ID |
Guca1a
guanylate cyclase activator 1a (retina) MGI:102770 |
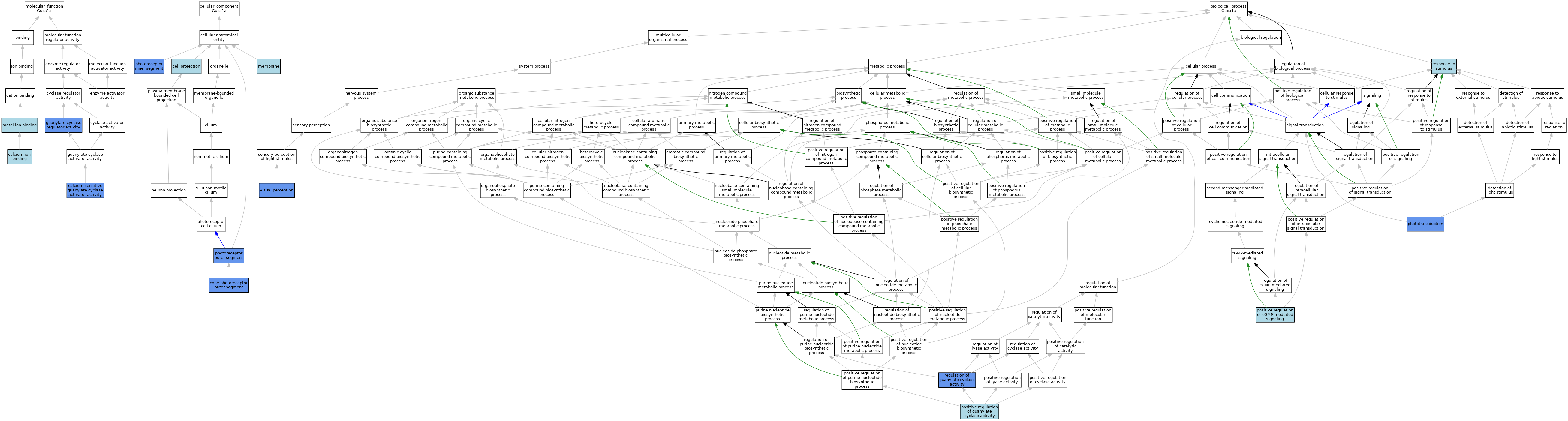
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008048 | calcium sensitive guanylate cyclase activator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008048 | calcium sensitive guanylate cyclase activator activity | IDA | J:83914 | |||||||||
| Molecular Function | GO:0030249 | guanylate cyclase regulator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030249 | guanylate cyclase regulator activity | IGI | J:92487 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0120199 | cone photoreceptor outer segment | ISO | J:164563 | |||||||||
| Cellular Component | GO:0120199 | cone photoreceptor outer segment | IDA | J:289584 | |||||||||
| Cellular Component | GO:0120199 | cone photoreceptor outer segment | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:289584 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | IDA | J:214071 | |||||||||
| Biological Process | GO:0007602 | phototransduction | IMP | J:99835 | |||||||||
| Biological Process | GO:0007602 | phototransduction | IMP | J:83914 | |||||||||
| Biological Process | GO:0010753 | positive regulation of cGMP-mediated signaling | ISO | J:155856 | |||||||||
| Biological Process | GO:0031284 | positive regulation of guanylate cyclase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0031284 | positive regulation of guanylate cyclase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0031282 | regulation of guanylate cyclase activity | IGI | J:92487 | |||||||||
| Biological Process | GO:0050896 | response to stimulus | IEA | J:60000 | |||||||||
| Biological Process | GO:0007601 | visual perception | IBA | J:265628 | |||||||||
| Biological Process | GO:0007601 | visual perception | IMP | J:99835 | |||||||||
| Biological Process | GO:0007601 | visual perception | IMP | J:83914 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||