|
Symbol Name ID |
Ddx4
DEAD box helicase 4 MGI:102670 |
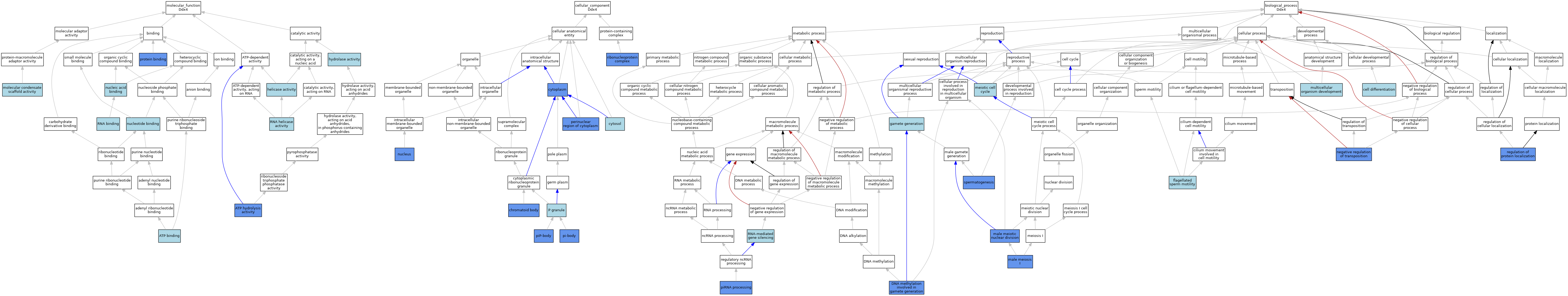
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IMP | J:243488 | |||||||||
| Molecular Function | GO:0004386 | helicase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0140693 | molecular condensate scaffold activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:189932 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:243444 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:148942 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107318 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:152071 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:87812 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:117063 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003724 | RNA helicase activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | ISO | J:62087 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | ISO | J:62087 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | ISO | J:62087 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | IDA | J:62087 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | IDA | J:189647 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | ISO | J:62087 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:96322 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:62087 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:83934 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:87735 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:62087 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:98391 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:62087 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5605205 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5605213 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5605279 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5605349 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5606226 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5606252 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5629217 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:83934 | |||||||||
| Cellular Component | GO:0043186 | P granule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IDA | J:62087 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IDA | J:179634 | |||||||||
| Cellular Component | GO:0071546 | pi-body | IDA | J:159518 | |||||||||
| Cellular Component | GO:0071546 | pi-body | IDA | J:155865 | |||||||||
| Cellular Component | GO:0071546 | pi-body | IDA | J:189647 | |||||||||
| Cellular Component | GO:0071547 | piP-body | IDA | J:155865 | |||||||||
| Cellular Component | GO:1990904 | ribonucleoprotein complex | IDA | J:117063 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0043046 | DNA methylation involved in gamete generation | IMP | J:159518 | |||||||||
| Biological Process | GO:0030317 | flagellated sperm motility | ISO | J:164563 | |||||||||
| Biological Process | GO:0007276 | gamete generation | IBA | J:265628 | |||||||||
| Biological Process | GO:0007141 | male meiosis I | IMP | J:243488 | |||||||||
| Biological Process | GO:0007141 | male meiosis I | IMP | J:61665 | |||||||||
| Biological Process | GO:0007140 | male meiotic nuclear division | IMP | J:159518 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0010529 | negative regulation of transposition | IMP | J:243488 | |||||||||
| Biological Process | GO:0034587 | piRNA processing | IMP | J:243488 | |||||||||
| Biological Process | GO:0034587 | piRNA processing | IMP | J:159518 | |||||||||
| Biological Process | GO:0032880 | regulation of protein localization | IMP | J:117063 | |||||||||
| Biological Process | GO:0031047 | RNA-mediated gene silencing | IEA | J:60000 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:159518 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:243488 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:61665 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||