|
Symbol Name ID |
Mcl1
myeloid cell leukemia sequence 1 MGI:101769 |
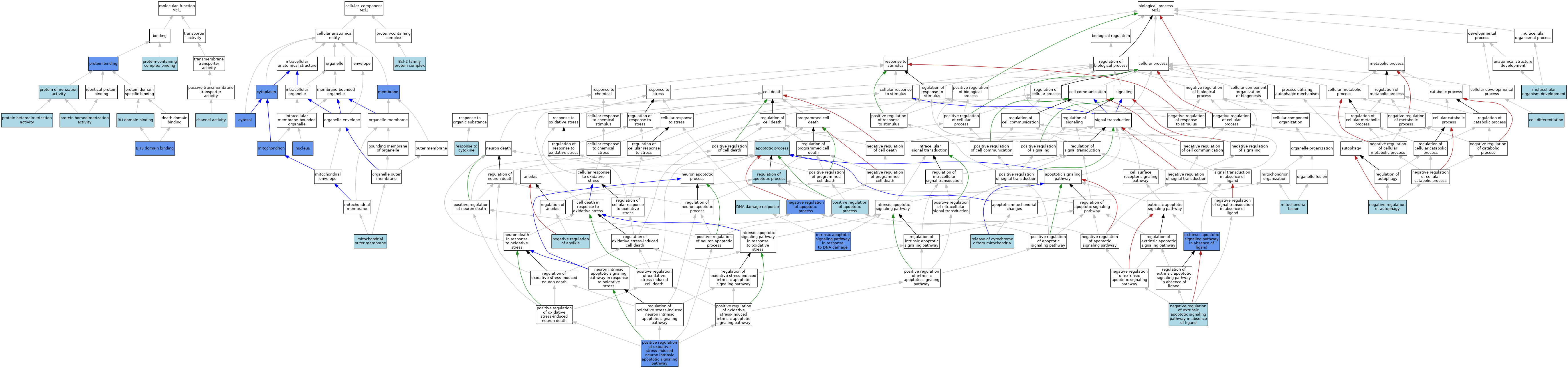
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051400 | BH domain binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051434 | BH3 domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051434 | BH3 domain binding | IPI | J:114727 | |||||||||
| Molecular Function | GO:0015267 | channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201566 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114705 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114727 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:163471 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182384 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182474 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:173142 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:235310 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:168877 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:203396 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:248256 | |||||||||
| Molecular Function | GO:0046983 | protein dimerization activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0097136 | Bcl-2 family protein complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097136 | Bcl-2 family protein complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:186356 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:201566 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:201566 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:23788 | |||||||||
| Cellular Component | GO:0005741 | mitochondrial outer membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:204747 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:23788 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:199864 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:186356 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:73065 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:73065 | |||||||||
| Biological Process | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | IMP | J:201566 | |||||||||
| Biological Process | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | ISO | J:164563 | |||||||||
| Biological Process | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | IBA | J:265628 | |||||||||
| Biological Process | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | IBA | J:265628 | |||||||||
| Biological Process | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | IDA | J:114727 | |||||||||
| Biological Process | GO:0008053 | mitochondrial fusion | IBA | J:265628 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:2000811 | negative regulation of anoikis | ISO | J:164563 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:182384 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010507 | negative regulation of autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | ISO | J:164563 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | IMP | J:204747 | |||||||||
| Biological Process | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0001836 | release of cytochrome c from mitochondria | IBA | J:265628 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | ISO | J:40789 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||