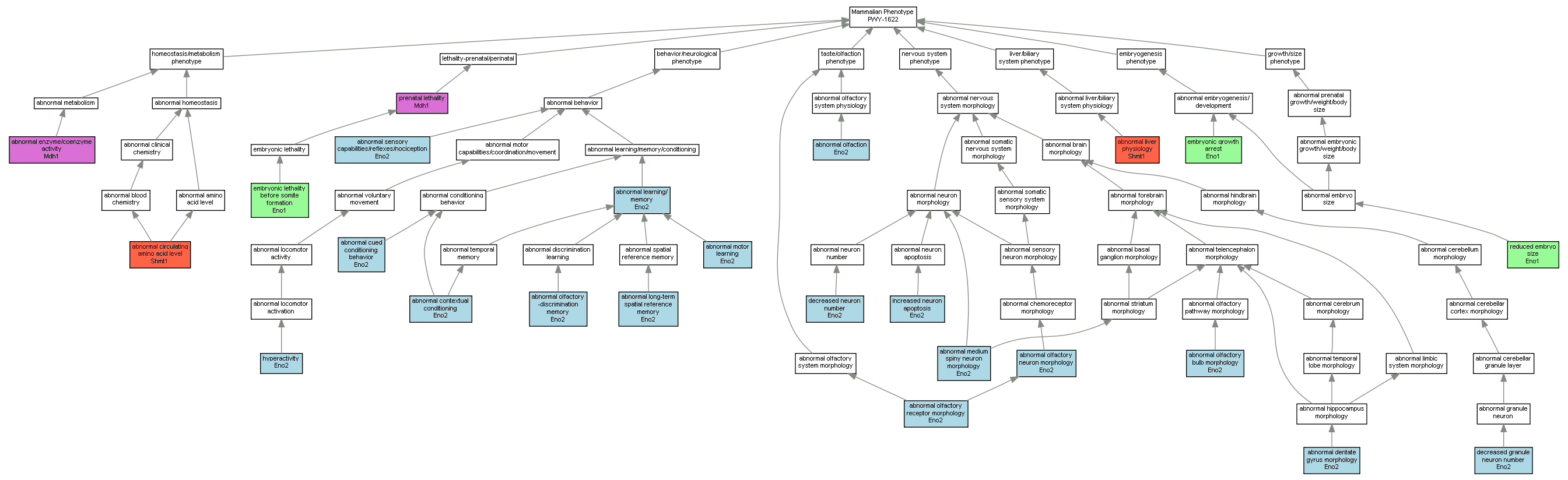
| ID | Term | Mouse gene |
Alleles | Background | Color Key |
| MP:0005311 | abnormal circulating amino acid level |
Shmt1 |
Shmt1tm1Stov/Shmt1+ | involves: 129S7/SvEvBrd * C57BL/6 | color key |
| MP:0005311 | abnormal circulating amino acid level |
Shmt1 |
Shmt1tm1Stov/Shmt1tm1Stov | involves: 129S7/SvEvBrd * C57BL/6 | color key |
| MP:0001469 | abnormal contextual conditioning |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0001454 | abnormal cued conditioning behavior |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0000812 | abnormal dentate gyrus morphology |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0005584 | abnormal enzyme/coenzyme activity |
Mdh1 |
Mdh1am1H/Mdh1+ | involves: C3H/HeH | color key |
| MP:0001449 | abnormal learning/ memory |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0000609 | abnormal liver physiology |
Shmt1 |
Shmt1tm1.1Stov/Shmt1tm1.1Stov | 129S/SvEv-Shmt1 | color key |
| MP:0000609 | abnormal liver physiology |
Shmt1 |
Shmt1tm1.1Stov/Shmt1tm1.1Stov | involves: 129S7/SvEvBrd * BALB/cJ * C57BL/6 | color key |
| MP:0000609 | abnormal liver physiology |
Shmt1 |
Shmt1tm1.1Stov/Shmt1+ | 129S/SvEv-Shmt1 | color key |
| MP:0008432 | abnormal long-term spatial reference memory |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0008462 | abnormal medium spiny neuron morphology |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0002804 | abnormal motor learning |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0001984 | abnormal olfaction |
Eno2 |
Acsm4tm1(cre)Hsak/?,Eno2tm1(DTA)Hsak/? | involves: 129S2/SvPas * C57BL/6 | color key |
| MP:0001460 | abnormal olfactory -discrimination memory |
Eno2 |
Acsm4tm1(cre)Hsak/?,Eno2tm1(DTA)Hsak/? | involves: 129S2/SvPas * C57BL/6 | color key |
| MP:0000819 | abnormal olfactory bulb morphology |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)5-1Kag/0 | involves: C57BL/6 | color key |
| MP:0000819 | abnormal olfactory bulb morphology |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0006092 | abnormal olfactory neuron morphology |
Eno2 |
Eno2tm1(DTA)Hsak/?,Tg(Olfr16-cre,-CFP)1Hsak/? | involves: C57BL/6 | color key |
| MP:0006092 | abnormal olfactory neuron morphology |
Eno2 |
Acsm4tm1(cre)Hsak/?,Eno2tm1(DTA)Hsak/? | involves: 129S2/SvPas * C57BL/6 | color key |
| MP:0001003 | abnormal olfactory receptor morphology |
Eno2 |
Eno2tm1(DTA)Hsak/?,Tg(Olfr16-cre,-CFP)1Hsak/? | involves: C57BL/6 | color key |
| MP:0001003 | abnormal olfactory receptor morphology |
Eno2 |
Acsm4tm1(cre)Hsak/?,Eno2tm1(DTA)Hsak/? | involves: 129S2/SvPas * C57BL/6 | color key |
| MP:0002067 | abnormal sensory capabilities/reflexes/nociception |
Eno2 |
Acsm4tm1(cre)Hsak/?,Eno2tm1(DTA)Hsak/? | involves: 129S2/SvPas * C57BL/6 | color key |
| MP:0002067 | abnormal sensory capabilities/reflexes/nociception |
Eno2 |
Eno2tm1(DTA)Hsak/?,Tg(Olfr16-cre,-CFP)1Hsak/? | involves: C57BL/6 | color key |
| MP:0008924 | decreased granule neuron number |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0008924 | decreased granule neuron number |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)5-1Kag/0 | involves: C57BL/6 | color key |
| MP:0008948 | decreased neuron number |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)4Kag/0 | involves: C57BL/6 | color key |
| MP:0008948 | decreased neuron number |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2tm1(DTA)Hsak,Tg(Nes-cre/ERT2)5-1Kag/0 | involves: C57BL/6 | color key |
| MP:0001730 | embryonic growth arrest |
Eno1 |
Eno1Gt(ROSABetageo)1Ev/Eno1Gt(ROSABetageo)1Ev | involves: 129S/SvEv | color key |
| MP:0006205 | embryonic lethality before somite formation |
Eno1 |
Eno1Gt(ROSABetageo)1Ev/Eno1Gt(ROSABetageo)1Ev | involves: 129S/SvEv | color key |
| MP:0001399 | hyperactivity |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0003203 | increased neuron apoptosis |
Eno2 |
Eno2tm1(DTA)Hsak/Eno2+,Gng7tm2(cre/PGR)Mmsh/Gng7+ | involves: C57BL/6 | color key |
| MP:0002080 | prenatal lethality |
Mdh1 |
Mdh1am1H/Mdh1am1H | involves: C3H/HeH | color key |
| MP:0001698 | reduced embryo size |
Eno1 |
Eno1Gt(ROSABetageo)1Ev/Eno1Gt(ROSABetageo)1Ev | involves: 129S/SvEv | color key |