How do I search for a gene using an Entrez, Ensembl, or other accession ID?
How do I search for a gene using an Entrez, Ensembl, or other accession ID?
|
| ||
| Overview | ||
| You can use Quick Search to find genes by function, disease, phenotype, identifier or other types of data in MGI. Quick Search returns a list of genome features, terms, or other search results that match your entry. | ||
| Accessing Quick Search | ||
|
You can access MGI Quick Search directly from the homepage, or at the upper right corner of most other MGI pages.
For this tutorial, open the MGI home page in a new window and locate Quick Search at the top center of the page to get started. |
|
|
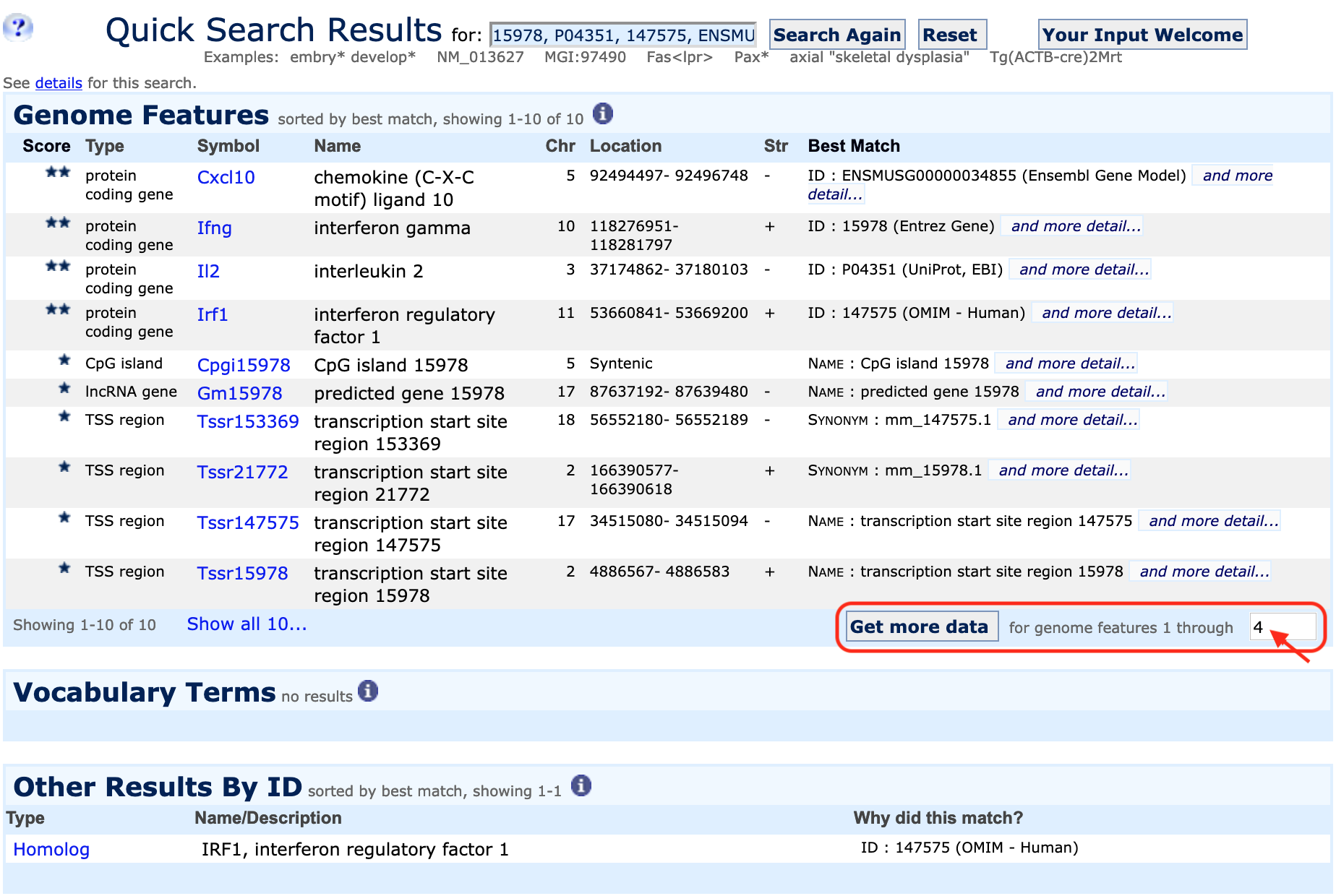
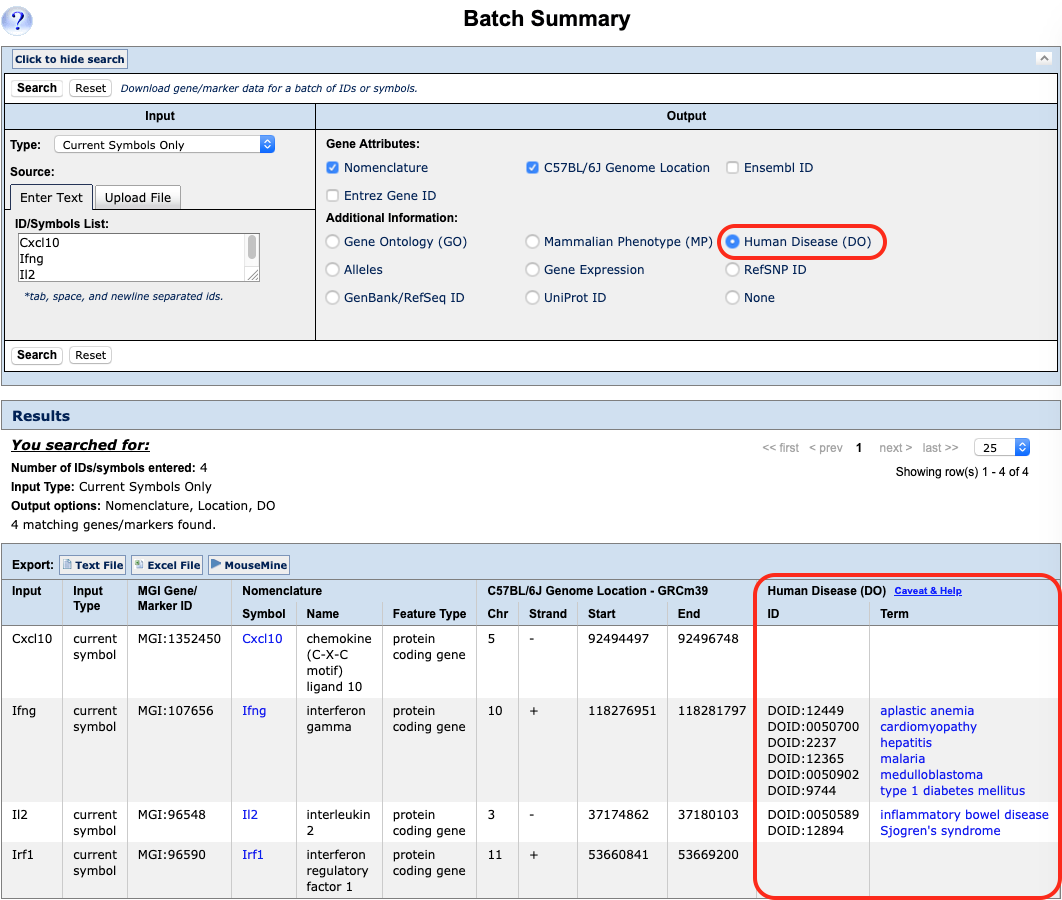
| Example 1. Using a list of gene symbols or identifiers as well as batch queries | |
|
|
|
|
|
You can also retrieve ontology (functional) information, alleles or expression data for all of your genes using this window. Results can be exported as plain text or Excel files. |
|
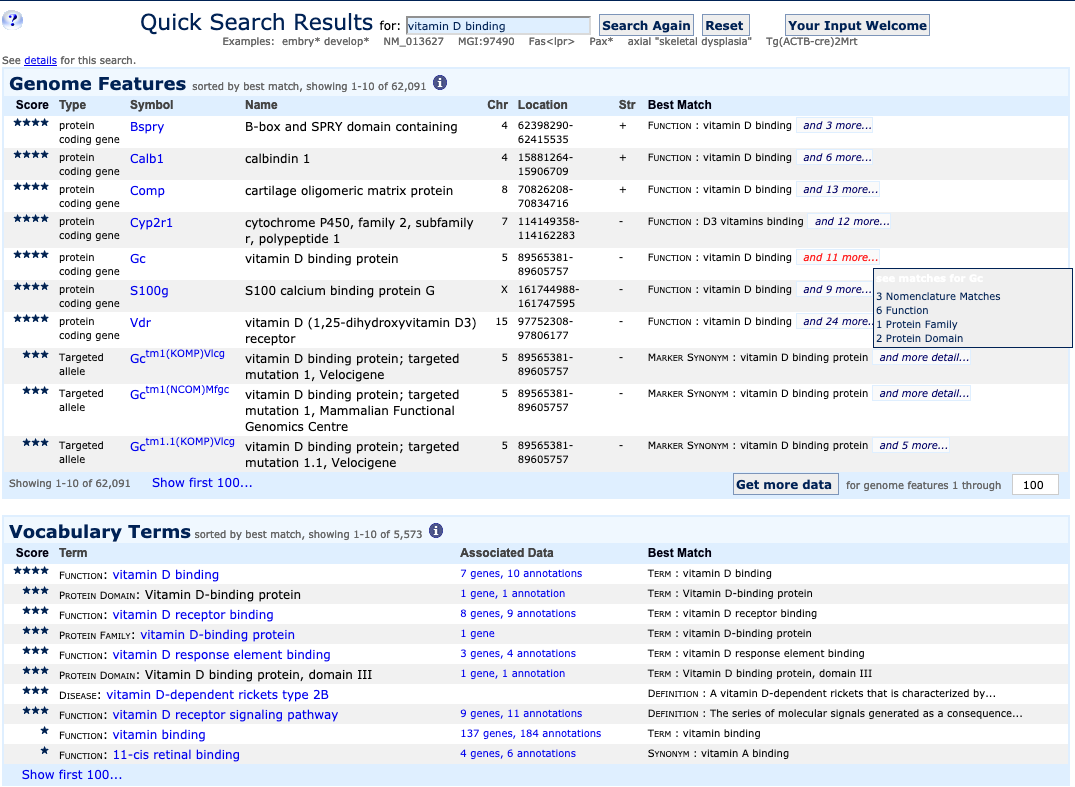
| Example 2. Search using phrases | ||
|
|
|
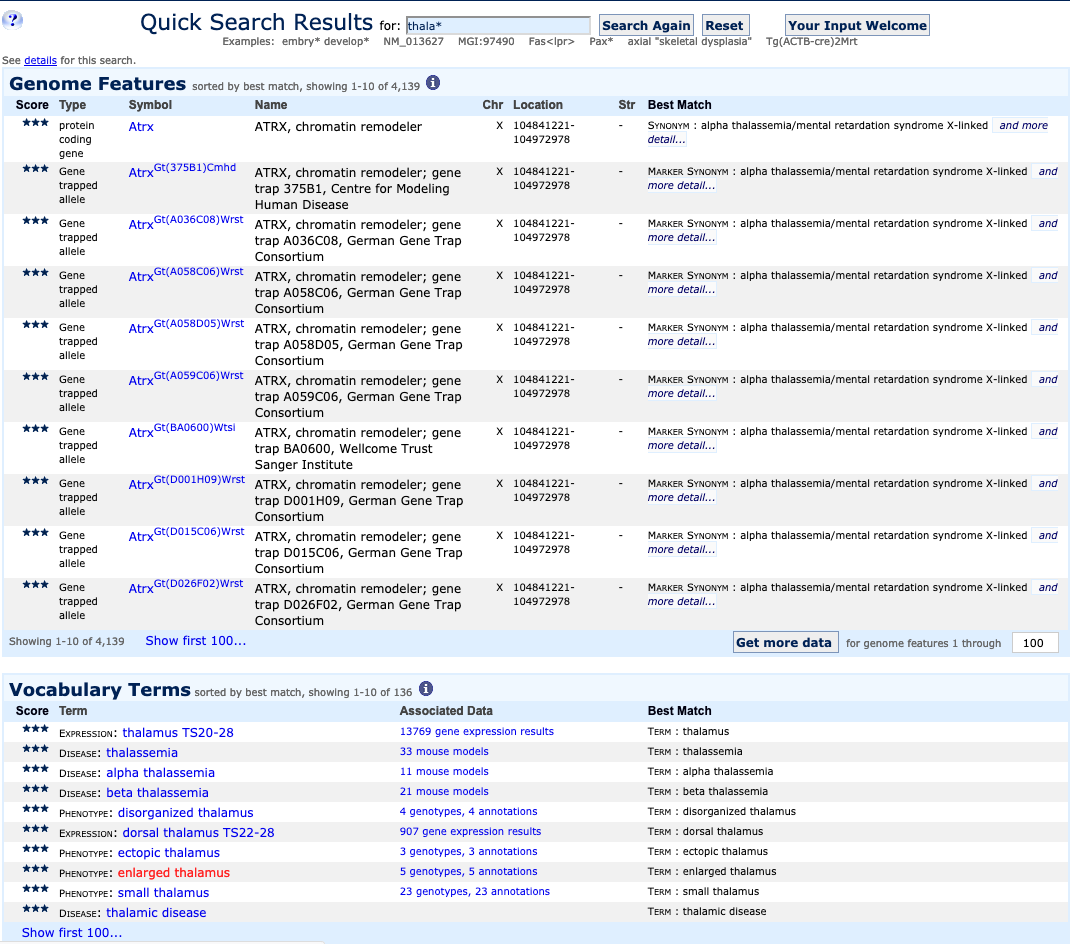
| Example 3. Expanding a search with wildcards | ||
|
|
|
|
|
|
| Example 4. Advanced searches using query forms | |
MGI query forms are the best option for answering very specific questions, especially those
that combine queries across major data content areas such as function and expression, phenotype
and genomic location, and so on. To view examples, click a FAQ link below.
|
|