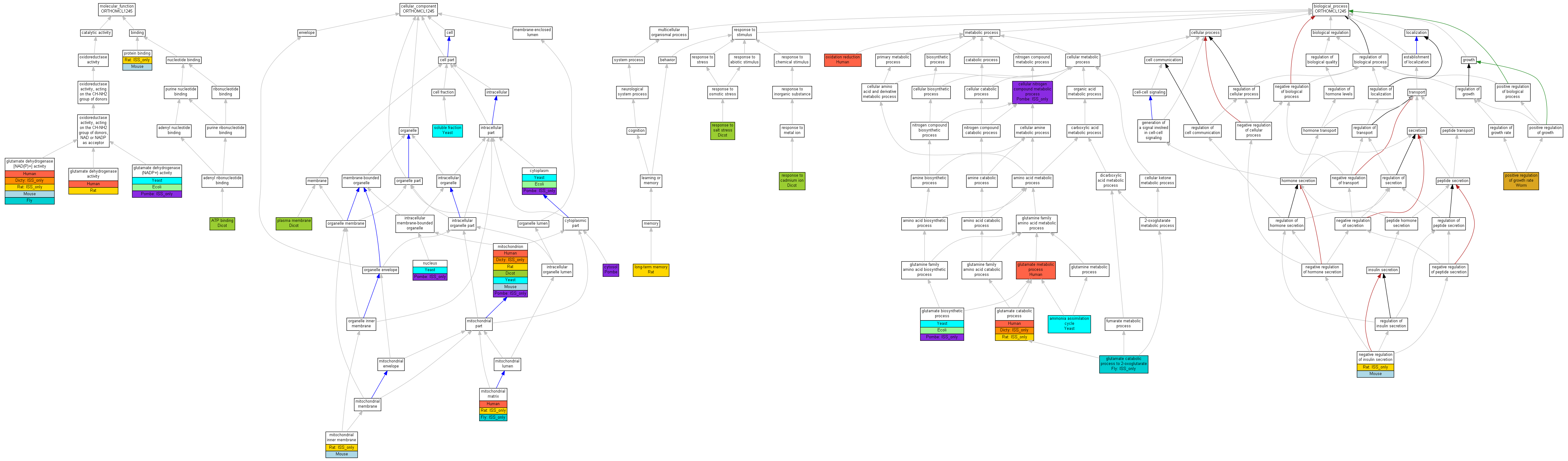
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0031975 | envelope | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0031974 | membrane-enclosed lumen | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0044422 | organelle part | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0051179 | localization | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | (Rat) | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | GDH2 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Molecular Function | GO:0005524 | ATP binding | GDH1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH1 | IDA | Yeast | SGD_REF:S000056962|PMID:2932370 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH1 | IDA | Yeast | SGD_REF:S000052640|PMID:2989290 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH1 | IMP | Yeast | SGD_REF:S000052640|PMID:2989290 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH1 | IDA | Yeast | SGD_REF:S000066220|PMID:11562373 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH1 | IMP | Yeast | SGD_REF:S000056962|PMID:2932370 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH3 | ISS | SGD:S000005902 | Yeast | SGD_REF:S000039624|PMID:7731988 |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH3 | IDA | Yeast | SGD_REF:S000066220|PMID:11562373 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | GDH3 | IMP | Yeast | SGD_REF:S000049665|PMID:9287019 | |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | SPCC622.12c | ISS | SGD:S000005902 | Pombe | PMID:17072883 |
| Molecular Function | GO:0004354 | glutamate dehydrogenase (NADP+) activity | gdhA | IDA | Ecoli | PMID:241744 | |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | P00367 | EXP | Human | PMID:429360 | |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Glud1 | IDA | Mouse | MGI:MGI:2157179|PMID:11792727 | |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Glud1 | IDA | Mouse | MGI:MGI:3769241|PMID:16959573 | |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Glud1 | ISO | RGD:1551567 | Rat | RGD:1624291 |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Glud1 | ISO | RGD:733213 | Rat | RGD:1624291 |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Gdh | IDA | Fly | FB:FBrf0063813 | |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Gdh | ISS | MGI:MGI:95753 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Gdh | ISS | UniProtKB:P00367 | Fly | FB:FBrf0159903 |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | Gdh | ISS | UniProt:P49448 | Fly | FB:FBrf0159903 |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity | glud1 | ISS | MGI:95753 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004352 | glutamate dehydrogenase activity | P49448 | IDA | Human | PMID:8207021 | |
| Molecular Function | GO:0004352 | glutamate dehydrogenase activity | P00367 | IDA | Human | PMID:15578726 | |
| Molecular Function | GO:0004352 | glutamate dehydrogenase activity | P49448 | IDA | Human | PMID:15578726 | |
| Molecular Function | GO:0004352 | glutamate dehydrogenase activity | Glud1 | IDA | Rat | RGD:1601356|PMID:15273247 | |
| Molecular Function | GO:0004352 | glutamate dehydrogenase activity | Glud1 | ISO | RGD:733213 | Rat | RGD:1624291 |
| Molecular Function | GO:0005515 | protein binding | Glud1 | IPI | UniProtKB:Q8R216 | Mouse | MGI:MGI:3769241|PMID:16959573 |
| Molecular Function | GO:0005515 | protein binding | Glud1 | ISO | RGD:1551567 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | GDH1 | IDA | Yeast | SGD_REF:S000069459|PMID:11914276 | |
| Cellular Component | GO:0005737 | cytoplasm | SPCC622.12c | ISS | SGD:S000005902 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005737 | cytoplasm | gdhA | IDA | Ecoli | PMID:241744 | |
| Cellular Component | GO:0005829 | cytosol | SPCC622.12c | IDA | Pombe | PMID:16823372 | |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Glud1 | IDA | Mouse | MGI:MGI:3590069|PMID:12865426 | |
| Cellular Component | GO:0005743 | mitochondrial inner membrane | Glud1 | ISO | RGD:1551567 | Rat | RGD:1624291 |
| Cellular Component | GO:0005759 | mitochondrial matrix | P00367 | EXP | Human | PMID:429360 | |
| Cellular Component | GO:0005759 | mitochondrial matrix | Glud1 | ISO | RGD:733213 | Rat | RGD:1624291 |
| Cellular Component | GO:0005759 | mitochondrial matrix | Gdh | ISS | Fly | FB:FBrf0141186 | |
| Cellular Component | GO:0005759 | mitochondrial matrix | Gdh | ISS | UniProtKB:P00367 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005759 | mitochondrial matrix | Gdh | ISS | UniProt:P49448 | Fly | FB:FBrf0159903 |
| Cellular Component | GO:0005739 | mitochondrion | P00367 | IDA | Human | PMID:15578726 | |
| Cellular Component | GO:0005739 | mitochondrion | Glud1 | IDA | Mouse | MGI:MGI:2682130|PMID:14651853 | |
| Cellular Component | GO:0005739 | mitochondrion | Glud1 | IDA | Mouse | MGI:MGI:3769241|PMID:16959573 | |
| Cellular Component | GO:0005739 | mitochondrion | Glud1 | ISO | RGD:733213 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | Glud1 | ISO | RGD:1551567 | Rat | RGD:1624291 |
| Cellular Component | GO:0005739 | mitochondrion | Glud1 | IDA | Rat | RGD:1601356|PMID:15273247 | |
| Cellular Component | GO:0005739 | mitochondrion | glud1 | ISS | MGI:95753 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005739 | mitochondrion | GDH2 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Cellular Component | GO:0005739 | mitochondrion | GDH1 | IDA | Dicot | TAIR:Publication:501720283|PMID:17137349 | |
| Cellular Component | GO:0005739 | mitochondrion | GDH2 | IDA | Dicot | TAIR:Publication:501724621|PMID:18385124 | |
| Cellular Component | GO:0005739 | mitochondrion | GDH1 | IDA | Dicot | TAIR:Publication:501724621|PMID:18385124 | |
| Cellular Component | GO:0005739 | mitochondrion | GDH3 | IDA | Yeast | SGD_REF:S000117178|PMID:16823961 | |
| Cellular Component | GO:0005739 | mitochondrion | GDH3 | IDA | Yeast | SGD_REF:S000075100|PMID:14576278 | |
| Cellular Component | GO:0005739 | mitochondrion | SPCC622.12c | ISS | SGD:S000000058 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005634 | nucleus | GDH1 | IDA | Yeast | SGD_REF:S000063234|PMID:4126 | |
| Cellular Component | GO:0005634 | nucleus | GDH3 | IDA | Yeast | SGD_REF:S000063234|PMID:4126 | |
| Cellular Component | GO:0005634 | nucleus | SPCC622.12c | ISS | SGD:S000005902 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005886 | plasma membrane | GDH2 | IDA | Dicot | TAIR:Publication:501720626|PMID:17151019 | |
| Cellular Component | GO:0005625 | soluble fraction | GDH1 | IDA | Yeast | SGD_REF:S000066220|PMID:11562373 | |
| Cellular Component | GO:0005625 | soluble fraction | GDH3 | IDA | Yeast | SGD_REF:S000066220|PMID:11562373 | |
| Biological Process | GO:0019676 | ammonia assimilation cycle | GDH1 | IEP | Yeast | SGD_REF:S000061424|PMID:11356843 | |
| Biological Process | GO:0019676 | ammonia assimilation cycle | GDH1 | IEP | Yeast | SGD_REF:S000042930|PMID:9611819 | |
| Biological Process | GO:0019676 | ammonia assimilation cycle | GDH3 | IEP | Yeast | SGD_REF:S000061424|PMID:11356843 | |
| Biological Process | GO:0034641 | cellular nitrogen compound metabolic process | SPCC622.12c | ISS | UniProtKB:P39708 | Pombe | PMID:17072883 |
| Biological Process | GO:0006537 | glutamate biosynthetic process | GDH1 | IGI | Yeast | SGD_REF:S000052640|PMID:2989290 | |
| Biological Process | GO:0006537 | glutamate biosynthetic process | GDH1 | IGI | Yeast | SGD_REF:S000056962|PMID:2932370 | |
| Biological Process | GO:0006537 | glutamate biosynthetic process | GDH3 | IMP | Yeast | SGD_REF:S000049665|PMID:9287019 | |
| Biological Process | GO:0006537 | glutamate biosynthetic process | GDH3 | IGI | Yeast | SGD_REF:S000049665|PMID:9287019 | |
| Biological Process | GO:0006537 | glutamate biosynthetic process | SPCC622.12c | ISS | SGD:S000005902 | Pombe | PMID:17072883 |
| Biological Process | GO:0006537 | glutamate biosynthetic process | gdhA | IDA | Ecoli | PMID:241744 | |
| Biological Process | GO:0006538 | glutamate catabolic process | P00367 | IDA | Human | PMID:6121377 | |
| Biological Process | GO:0006538 | glutamate catabolic process | Glud1 | ISO | RGD:733213 | Rat | RGD:1624291 |
| Biological Process | GO:0006538 | glutamate catabolic process | glud1 | ISS | UniProt:P00367 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | Gdh | ISS | UniProtKB:P00367 | Fly | FB:FBrf0159903 |
| Biological Process | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | Gdh | ISS | UniProt:P49448 | Fly | FB:FBrf0159903 |
| Biological Process | GO:0006536 | glutamate metabolic process | P49448 | IDA | Human | PMID:8207021 | |
| Biological Process | GO:0007616 | long-term memory | Glud1 | IEP | Rat | RGD:632875|PMID:9275181 | |
| Biological Process | GO:0046676 | negative regulation of insulin secretion | Glud1 | IMP | Mouse | MGI:MGI:3769241|PMID:16959573 | |
| Biological Process | GO:0046676 | negative regulation of insulin secretion | Glud1 | IGI | MGI:MGI:1922637 | Mouse | MGI:MGI:3769241|PMID:16959573 |
| Biological Process | GO:0046676 | negative regulation of insulin secretion | Glud1 | ISO | RGD:1551567 | Rat | RGD:1624291 |
| Biological Process | GO:0055114 | oxidation reduction | P49448 | IDA | Human | PMID:8207021 | |
| Biological Process | GO:0040010 | positive regulation of growth rate | ZK829.4 | IMP | WB:WBRNAi00038456|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0046686 | response to cadmium ion | GDH2 | IEP | Dicot | TAIR:Publication:501718481|PMID:16502469 | |
| Biological Process | GO:0046686 | response to cadmium ion | GDH1 | IEP | Dicot | TAIR:Publication:501718481|PMID:16502469 | |
| Biological Process | GO:0009651 | response to salt stress | GDH2 | IEP | Dicot | TAIR:Publication:501723430|PMID:17916636 | |
| Biological Process | GO:0009651 | response to salt stress | GDH1 | IEP | Dicot | TAIR:Publication:501723430|PMID:17916636 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |