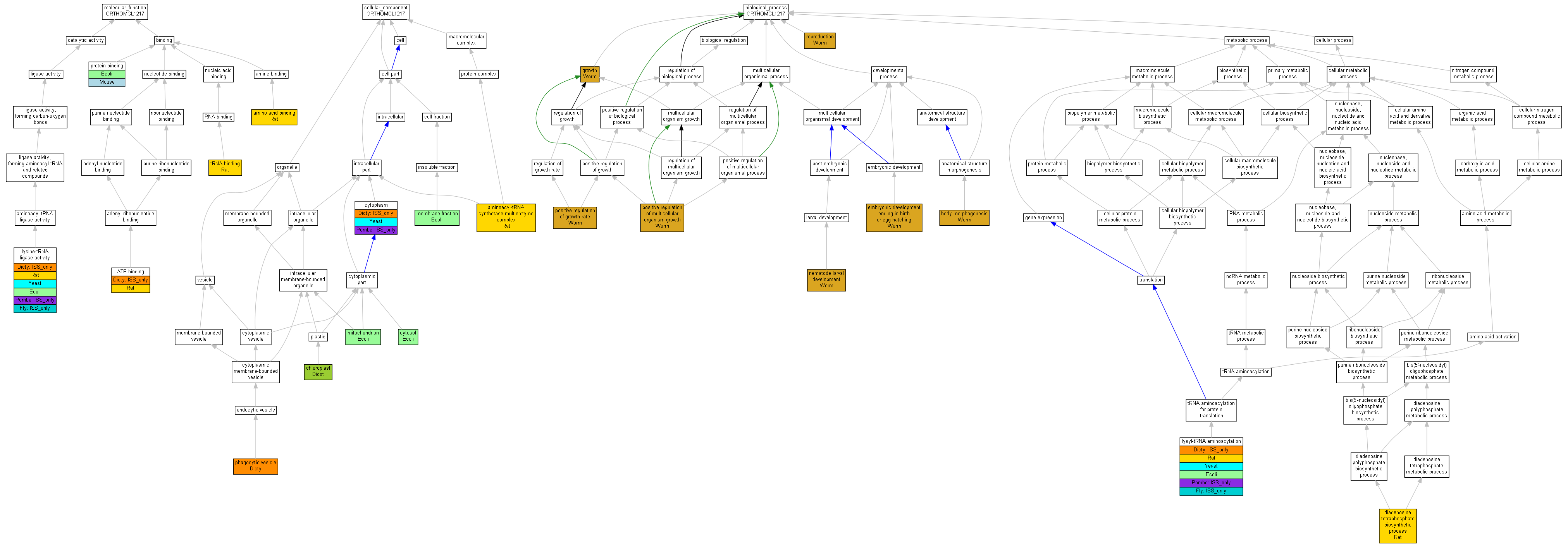
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | (Dicty) | Dicot | Yeast | Pombe | Ecoli |
| Molecular Function | GO:0003824 | catalytic activity | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | (Pombe) | Ecoli |
| Cellular Component | GO:0032991 | macromolecular complex | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Cellular Component | GO:0043226 | organelle | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0040007 | growth | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | (Fly) | Worm | (Dicty) | Dicot | Yeast | (Pombe) | Ecoli |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0048518 | positive regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Biological Process | GO:0000003 | reproduction | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016597 | amino acid binding | Kars | IDA | Rat | RGD:2303396|PMID:3988754 | |
| Molecular Function | GO:0016597 | amino acid binding | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Molecular Function | GO:0016597 | amino acid binding | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Molecular Function | GO:0005524 | ATP binding | Kars | IDA | Rat | RGD:2303396|PMID:3988754 | |
| Molecular Function | GO:0005524 | ATP binding | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Molecular Function | GO:0005524 | ATP binding | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Molecular Function | GO:0005524 | ATP binding | lysS | ISS | InterPro:IPR002313 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | Aats-lys | ISS | SGD:S0002444 | Fly | FB:FBrf0105495 |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | lysS | ISS | SGD:S000002444 | Dicty | dictyBase_REF:10155 |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | KRS1 | IDA | Yeast | SGD_REF:S000126709|PMID:3888626 | |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | KRS1 | IMP | Yeast | SGD_REF:S000063840|PMID:7628447 | |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | krs1 | ISS | UniProtKB:P15180 | Pombe | PMID:17072883 |
| Molecular Function | GO:0004824 | lysine-tRNA ligase activity | lysU | IDA | Ecoli | PMID:7782306 | |
| Molecular Function | GO:0005515 | protein binding | Kars | IPI | UniProtKB:P25118 | Mouse | MGI:MGI:3694174|PMID:16291755 |
| Molecular Function | GO:0005515 | protein binding | lysS | IPI | UniProtKB:P43672 | Ecoli | PMID:15690043 |
| Molecular Function | GO:0000049 | tRNA binding | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Molecular Function | GO:0000049 | tRNA binding | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Cellular Component | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Cellular Component | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex | Kars | IDA | Rat | RGD:2303396|PMID:3988754 | |
| Cellular Component | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Cellular Component | GO:0009507 | chloroplast | OVA5 | IDA | Dicot | TAIR:Publication:501724486|PMID:18431481 | |
| Cellular Component | GO:0005737 | cytoplasm | lysS | ISS | SGD:S000002444 | Dicty | dictyBase_REF:10155 |
| Cellular Component | GO:0005737 | cytoplasm | KRS1 | IDA | Yeast | SGD_REF:S000063837|PMID:330225 | |
| Cellular Component | GO:0005737 | cytoplasm | krs1 | ISS | UniProtKB:P15180 | Pombe | PMID:17072883 |
| Cellular Component | GO:0005829 | cytosol | lysS | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005829 | cytosol | lysU | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005624 | membrane fraction | lysS | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005624 | membrane fraction | lysU | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005739 | mitochondrion | lysS | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0005739 | mitochondrion | lysU | IDA | Ecoli | PMID:16858726 | |
| Cellular Component | GO:0045335 | phagocytic vesicle | lysS | IDA | Dicty | dictyBase_REF:11413|PMID:16926386 | |
| Biological Process | GO:0010171 | body morphogenesis | krs-1 | IMP | WB:WBRNAi00009101|WB:WBPhenotype:0000535 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0015966 | diadenosine tetraphosphate biosynthetic process | Kars | IDA | Rat | RGD:2303395|PMID:6626505 | |
| Biological Process | GO:0015966 | diadenosine tetraphosphate biosynthetic process | Kars | IDA | Rat | RGD:2303396|PMID:3988754 | |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | krs-1 | IMP | WB:WBRNAi00009101|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0009792 | embryonic development ending in birth or egg hatching | krs-1 | IMP | WB:WBRNAi00026167|WB:WBPhenotype:0000050 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0040007 | growth | krs-1 | IMP | WB:WBRNAi00026167|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0040007 | growth | krs-1 | IMP | WB:WBRNAi00035072|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | Kars | IDA | Rat | RGD:2303393|PMID:7372681 | |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | Kars | IDA | Rat | RGD:2303394|PMID:7082655 | |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | Aats-lys | ISS | SGD:S0002444 | Fly | FB:FBrf0105495 |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | lysS | ISS | SGD:S000002444 | Dicty | dictyBase_REF:10155 |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | KRS1 | IMP | Yeast | SGD_REF:S000063840|PMID:7628447 | |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | KRS1 | IDA | Yeast | SGD_REF:S000126709|PMID:3888626 | |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | krs1 | ISS | UniProtKB:P15180 | Pombe | PMID:17072883 |
| Biological Process | GO:0006430 | lysyl-tRNA aminoacylation | lysU | IDA | Ecoli | PMID:7782306 | |
| Biological Process | GO:0002119 | nematode larval development | krs-1 | IMP | WB:WBRNAi00026167|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00006395|PMID:14551910 |
| Biological Process | GO:0002119 | nematode larval development | krs-1 | IMP | WB:WBRNAi00035072|WB:WBPhenotype:0000059 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0040010 | positive regulation of growth rate | krs-1 | IMP | WB:WBRNAi00009101|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0040010 | positive regulation of growth rate | krs-1 | IMP | WB:WBRNAi00035072|WB:WBPhenotype:0000031 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | krs-1 | IMP | WB:WBRNAi00009101|WB:WBPhenotype:0000229 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | krs-1 | IMP | WB:WBRNAi00009101|WB:WBPhenotype:0001037 | Worm | WB:WBPaper00005654|PMID:12529635 |
| Biological Process | GO:0000003 | reproduction | krs-1 | IMP | WB:WBRNAi00035072|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00024497|PMID:15489339 |
| Biological Process | GO:0000003 | reproduction | krs-1 | IMP | WB:WBRNAi00052232|WB:WBPhenotype:0000689 | Worm | WB:WBPaper00025054|PMID:15791247 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |