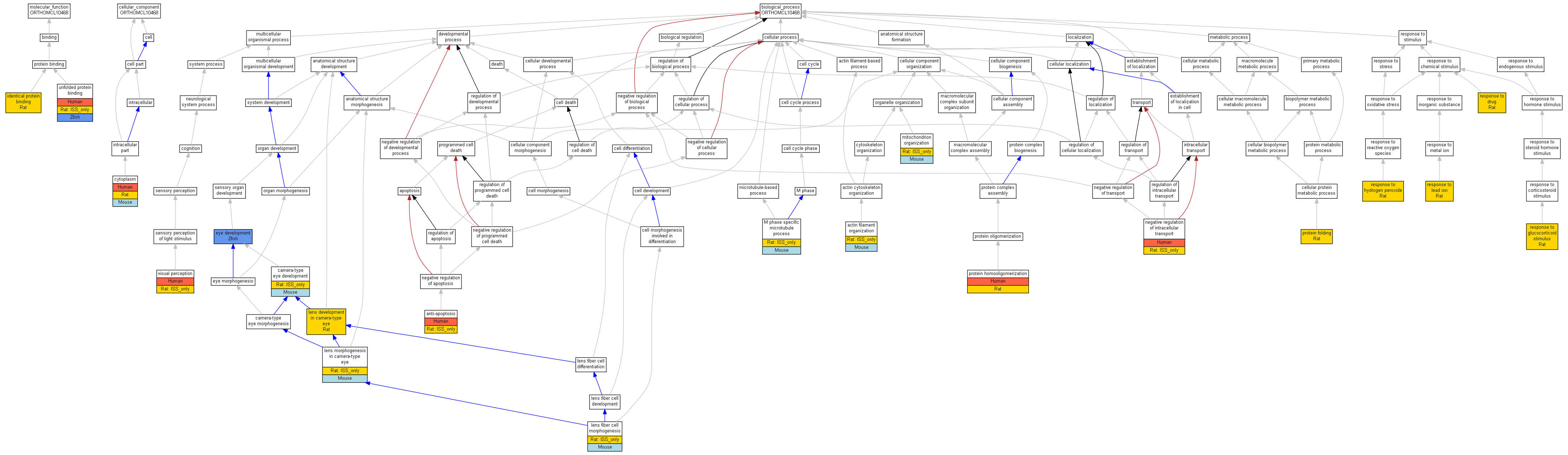
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0005623 | cell | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Cellular Component | GO:0044464 | cell part | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0010926 | anatomical structure formation | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0065007 | biological regulation | Human | Mouse | (Rat) | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0009987 | cellular process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0032502 | developmental process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0051234 | establishment of localization | Human | Mouse | (Rat) | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0051179 | localization | Human | Mouse | (Rat) | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0008152 | metabolic process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | Human | Mouse | (Rat) | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Biological Process | GO:0050896 | response to stimulus | Human | Mouse | Rat | Chicken | Zfish | Fly | X | X | X | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | Cryaa | IDA | Rat | RGD:1600986|PMID:14529298 | |
| Molecular Function | GO:0051082 | unfolded protein binding | P02489 | IPI | UniProtKB:P07320 | Human | PMID:8943244 |
| Molecular Function | GO:0051082 | unfolded protein binding | P02489 | IMP | Human | PMID:18407550 | |
| Molecular Function | GO:0051082 | unfolded protein binding | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| Molecular Function | GO:0051082 | unfolded protein binding | cryaa | IDA | Zfish | ZFIN:ZDB-PUB-050209-6|PMID:15692462 | |
| Cellular Component | GO:0005737 | cytoplasm | P02489 | IDA | Human | PMID:14752512 | |
| Cellular Component | GO:0005737 | cytoplasm | Cryaa | IDA | Mouse | MGI:MGI:2653777|PMID:12584250 | |
| Cellular Component | GO:0005737 | cytoplasm | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Cellular Component | GO:0005737 | cytoplasm | Cryaa | IDA | Rat | RGD:1600987|PMID:11851352 | |
| Biological Process | GO:0007015 | actin filament organization | Cryaa | IMP | MGI:MGI:3665275 | Mouse | MGI:MGI:3655912|PMID:16799046 |
| Biological Process | GO:0007015 | actin filament organization | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0006916 | anti-apoptosis | P02489 | IDA | Human | PMID:14752512 | |
| Biological Process | GO:0006916 | anti-apoptosis | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| Biological Process | GO:0043010 | camera-type eye development | Cryaa | IGI | MGI:MGI:1916503 | Mouse | MGI:MGI:2682441|PMID:12546709 |
| Biological Process | GO:0043010 | camera-type eye development | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0001654 | eye development | cryaa | IDA | Zfish | ZFIN:ZDB-PUB-060616-7|PMID:16728471 | |
| Biological Process | GO:0002088 | lens development in camera-type eye | Cryaa | IEP | Rat | RGD:1600991|PMID:8187157 | |
| Biological Process | GO:0070309 | lens fiber cell morphogenesis | Cryaa | IMP | MGI:MGI:3665275 | Mouse | MGI:MGI:3655912|PMID:16799046 |
| Biological Process | GO:0070309 | lens fiber cell morphogenesis | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0002089 | lens morphogenesis in camera-type eye | Cryaa | IMP | MGI:MGI:1858023 | Mouse | MGI:MGI:82388|PMID:8812430 |
| Biological Process | GO:0002089 | lens morphogenesis in camera-type eye | Cryaa | IMP | MGI:MGI:3665275|MGI:MGI:3665274 | Mouse | MGI:MGI:3655912|PMID:16799046 |
| Biological Process | GO:0002089 | lens morphogenesis in camera-type eye | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0000072 | M phase specific microtubule process | Cryaa | IMP | MGI:MGI:1858025 | Mouse | MGI:MGI:2653777|PMID:12584250 |
| Biological Process | GO:0000072 | M phase specific microtubule process | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0007005 | mitochondrion organization | Cryaa | IMP | MGI:MGI:3665275 | Mouse | MGI:MGI:3655912|PMID:16799046 |
| Biological Process | GO:0007005 | mitochondrion organization | Cryaa | ISO | RGD:10399 | Rat | RGD:1624291 |
| Biological Process | GO:0032387 | negative regulation of intracellular transport | P02489 | IDA | Human | PMID:14752512 | |
| Biological Process | GO:0032387 | negative regulation of intracellular transport | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| Biological Process | GO:0006457 | protein folding | Cryaa | IDA | Rat | RGD:1600989|PMID:11598124 | |
| Biological Process | GO:0051260 | protein homooligomerization | P02489 | IDA | Human | PMID:16303126 | |
| Biological Process | GO:0051260 | protein homooligomerization | Cryaa | IDA | Rat | RGD:1600986|PMID:14529298 | |
| Biological Process | GO:0051260 | protein homooligomerization | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| Biological Process | GO:0042493 | response to drug | Cryaa | IEP | Rat | RGD:1600987|PMID:11851352 | |
| Biological Process | GO:0051384 | response to glucocorticoid stimulus | Cryaa | IEP | Rat | RGD:2303613|PMID:18626730 | |
| Biological Process | GO:0042542 | response to hydrogen peroxide | Cryaa | IDA | Rat | RGD:1600990|PMID:7556464 | |
| Biological Process | GO:0010288 | response to lead ion | Cryaa | IEP | Rat | RGD:1600983|PMID:15905016 | |
| Biological Process | GO:0007601 | visual perception | P02489 | IMP | Human | PMID:14512969 | |
| Biological Process | GO:0007601 | visual perception | P02489 | IMP | Human | PMID:18587492 | |
| Biological Process | GO:0007601 | visual perception | P02489 | IMP | Human | PMID:9467006 | |
| Biological Process | GO:0007601 | visual perception | Cryaa | ISO | RGD:735272 | Rat | RGD:1624291 |
| EXP Inferred from experiment |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |