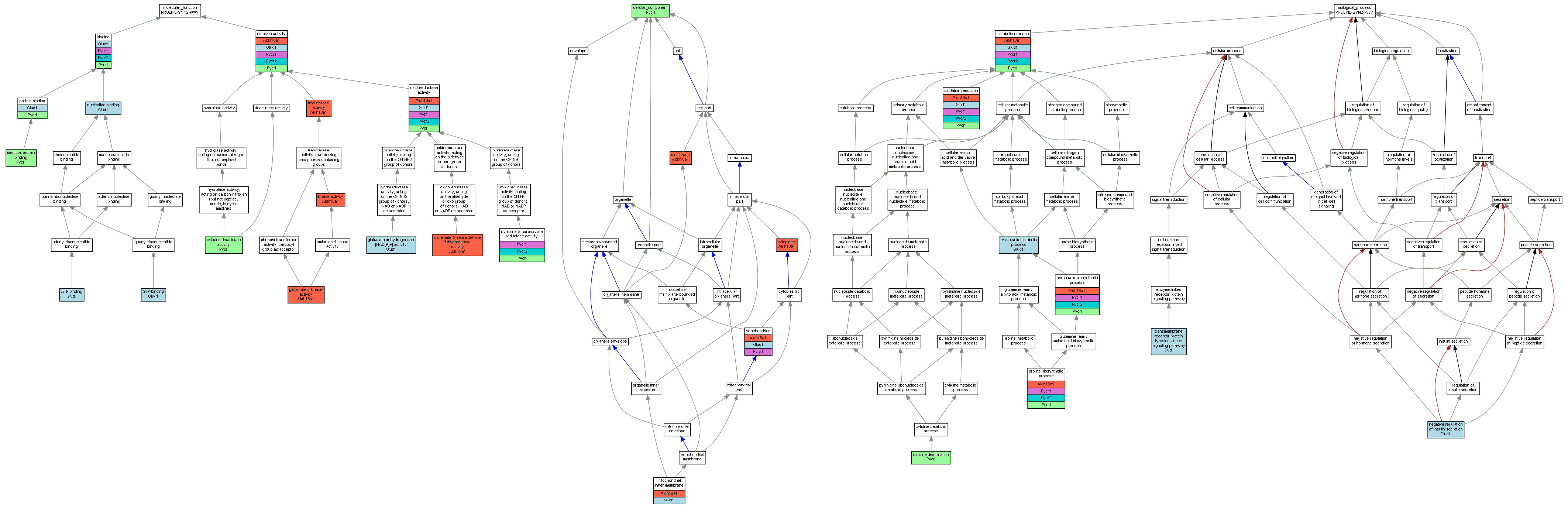
| Category | ID | Term | Mouse gene |
Evidence | Color Key |
| Molecular Function | GO:0005524 | ATP binding |
Glud1 |
IEA | color key |
| Molecular Function | GO:0005488 | binding |
Glud1 |
IEA | color key |
| Molecular Function | GO:0005488 | binding |
Pycr1 |
IEA | color key |
| Molecular Function | GO:0005488 | binding |
Pycr2 |
IEA | color key |
| Molecular Function | GO:0005488 | binding |
Pycrl |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Aldh18a1 |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Glud1 |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Pycr1 |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Pycr2 |
IEA | color key |
| Molecular Function | GO:0003824 | catalytic activity |
Pycrl |
IEA | color key |
| Molecular Function | GO:0004126 | cytidine deaminase activity |
Pycrl |
ISO | color key |
| Molecular Function | GO:0004349 | glutamate 5-kinase activity |
Aldh18a1 |
IDA | color key |
| Molecular Function | GO:0004349 | glutamate 5-kinase activity |
Aldh18a1 |
IGI | color key |
| Molecular Function | GO:0004353 | glutamate dehydrogenase [NAD(P)+] activity |
Glud1 |
IDA | color key |
| Molecular Function | GO:0004350 | glutamate-5-semialdehyde dehydrogenase activity |
Aldh18a1 |
IEA | color key |
| Molecular Function | GO:0005525 | GTP binding |
Glud1 |
IEA | color key |
| Molecular Function | GO:0042802 | identical protein binding |
Pycrl |
IPI | color key |
| Molecular Function | GO:0016301 | kinase activity |
Aldh18a1 |
IEA | color key |
| Molecular Function | GO:0000166 | nucleotide binding |
Glud1 |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Aldh18a1 |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Glud1 |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Pycr1 |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Pycr2 |
IEA | color key |
| Molecular Function | GO:0016491 | oxidoreductase activity |
Pycrl |
IEA | color key |
| Molecular Function | GO:0005515 | protein binding |
Glud1 |
IPI | color key |
| Molecular Function | GO:0005515 | protein binding |
Pycrl |
IPI | color key |
| Molecular Function | GO:0004735 | pyrroline-5-carboxylate reductase activity |
Pycr1 |
IEA | color key |
| Molecular Function | GO:0004735 | pyrroline-5-carboxylate reductase activity |
Pycr2 |
IEA | color key |
| Molecular Function | GO:0004735 | pyrroline-5-carboxylate reductase activity |
Pycrl |
IEA | color key |
| Molecular Function | GO:0016740 | transferase activity |
Aldh18a1 |
IEA | color key |
| Cellular Component | GO:0005575 | cellular_component |
Pycrl |
ND | color key |
| Cellular Component | GO:0005737 | cytoplasm |
Aldh18a1 |
IEA | color key |
| Cellular Component | GO:0016020 | membrane |
Aldh18a1 |
IEA | color key |
| Cellular Component | GO:0005743 | mitochondrial inner membrane |
Aldh18a1 |
IEA | color key |
| Cellular Component | GO:0005743 | mitochondrial inner membrane |
Glud1 |
IDA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Aldh18a1 |
IDA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Glud1 |
IDA | color key |
| Cellular Component | GO:0005739 | mitochondrion |
Pycr1 |
IDA | color key |
| Biological Process | GO:0008652 | amino acid biosynthetic process |
Aldh18a1 |
TAS | color key |
| Biological Process | GO:0008652 | amino acid biosynthetic process |
Pycr1 |
IEA | color key |
| Biological Process | GO:0008652 | amino acid biosynthetic process |
Pycr2 |
IEA | color key |
| Biological Process | GO:0008652 | amino acid biosynthetic process |
Pycrl |
IEA | color key |
| Biological Process | GO:0006520 | amino acid metabolic process |
Glud1 |
IEA | color key |
| Biological Process | GO:0009972 | cytidine deamination |
Pycrl |
ISO | color key |
| Biological Process | GO:0008152 | metabolic process |
Aldh18a1 |
IEA | color key |
| Biological Process | GO:0008152 | metabolic process |
Glud1 |
IEA | color key |
| Biological Process | GO:0008152 | metabolic process |
Pycr1 |
IEA | color key |
| Biological Process | GO:0008152 | metabolic process |
Pycr2 |
IEA | color key |
| Biological Process | GO:0008152 | metabolic process |
Pycrl |
IEA | color key |
| Biological Process | GO:0046676 | negative regulation of insulin secretion |
Glud1 |
IGI | color key |
| Biological Process | GO:0046676 | negative regulation of insulin secretion |
Glud1 |
IMP | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Aldh18a1 |
IEA | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Glud1 |
IEA | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Pycr1 |
IEA | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Pycr2 |
IEA | color key |
| Biological Process | GO:0055114 | oxidation reduction |
Pycrl |
IEA | color key |
| Biological Process | GO:0006561 | proline biosynthetic process |
Aldh18a1 |
IEA | color key |
| Biological Process | GO:0006561 | proline biosynthetic process |
Pycr1 |
IEA | color key |
| Biological Process | GO:0006561 | proline biosynthetic process |
Pycr2 |
IEA | color key |
| Biological Process | GO:0006561 | proline biosynthetic process |
Pycrl |
IEA | color key |
| Biological Process | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway |
Glud1 |
NAS | color key |