How do I search for genes by expression profile?
How do I search for genes by expression profile?
| Overview | ||
|
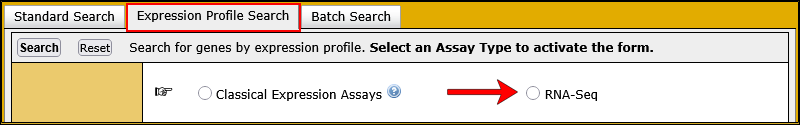
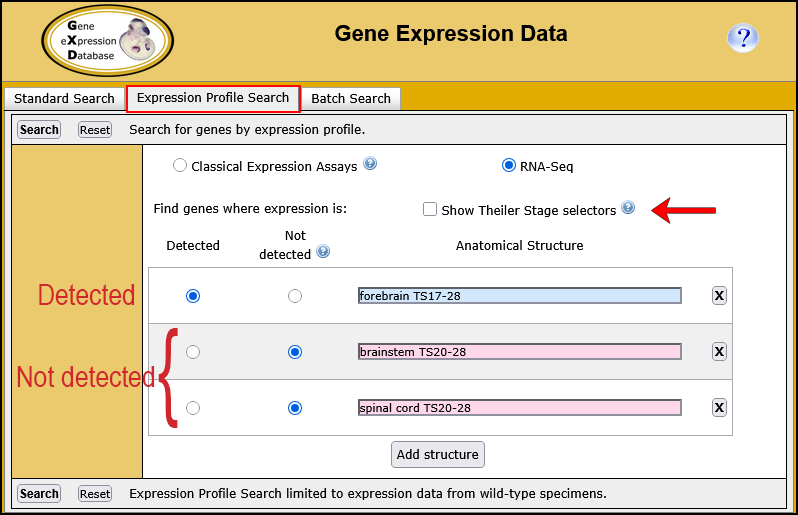
You can use the Expression Profile Search to define the expression profile of interest by specifying up to 10 anatomical structures and whether expression is present or absent in these structures as determined by either classical expression assays or RNA-Seq experiments. In addition, you can also specify the developmental (Theiler) stage(s) of interest.
|
||
| Accessing the Expression Profile Search | ||
|
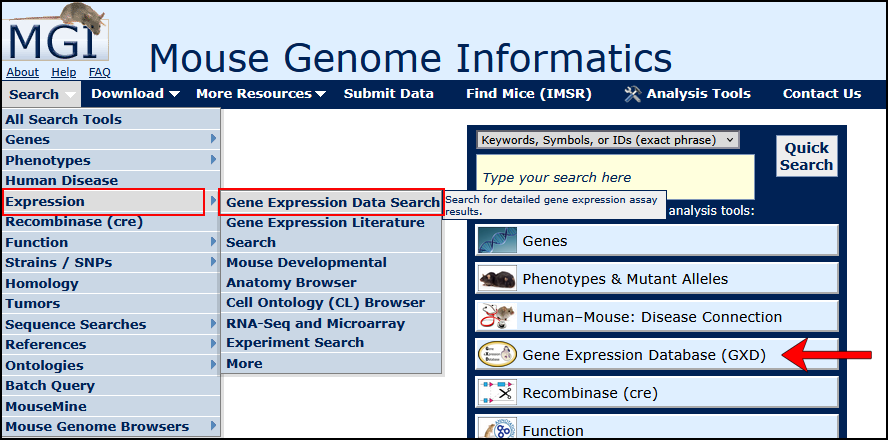
The Expression Profile Search is a tab on the Gene Expression Data Search.
On most MGI pages, you can access the Gene Expression Data Search from the Search menu as shown in the image at right.
Alternatively, you can also navigate to the Gene Expression Database (GXD) home page using the tile found on the MGI home page (arrow in image at right). Once there, click on the Expression Profile Search tile. For this tutorial, open the Gene Expression Data Search Form in a new window. Scroll down this page for further instructions. |
|
|
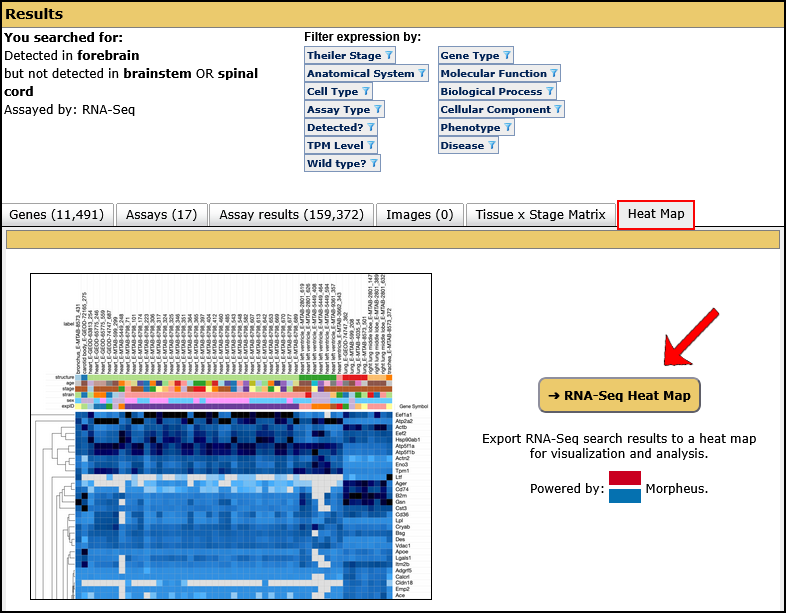
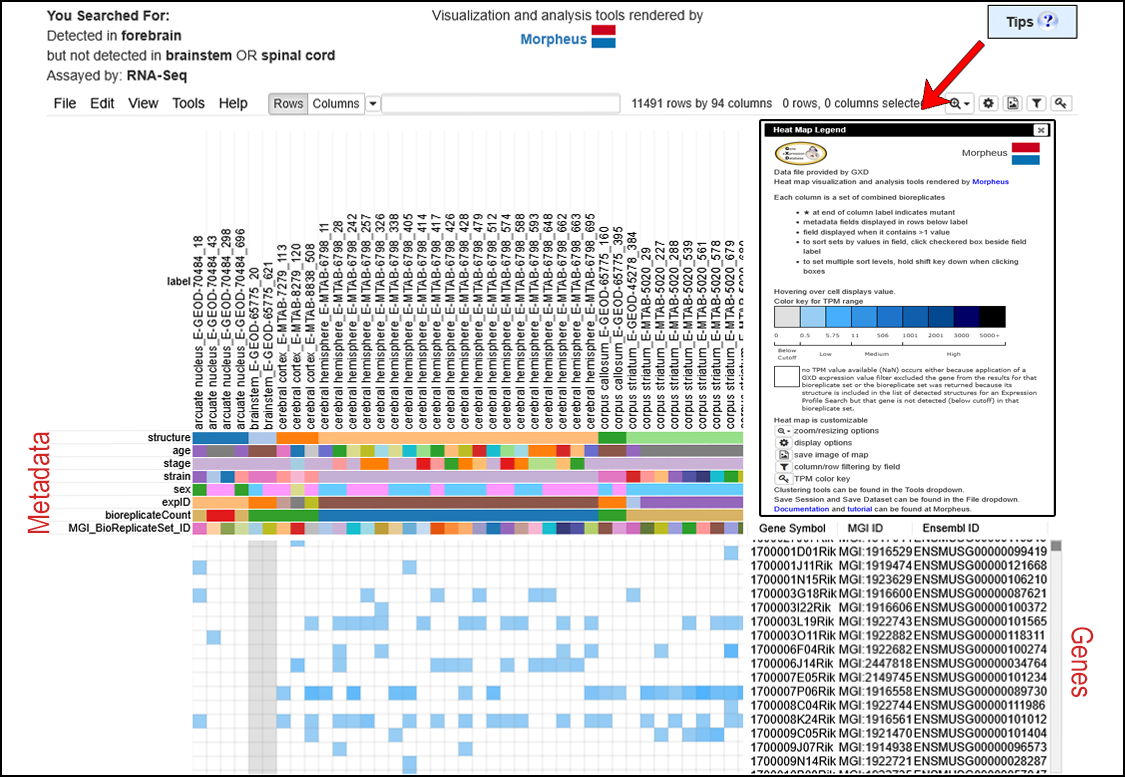
| Example. Search for genes expressed in the forebrain but not in the hindbrain or spinal cord as determined by RNA-Seq. | ||
|
|
|
|
|
|
|
|
|
|
|
|