How do I find expression data for a cell type?
How do I find expression data for a cell type?
| Overview | ||
|
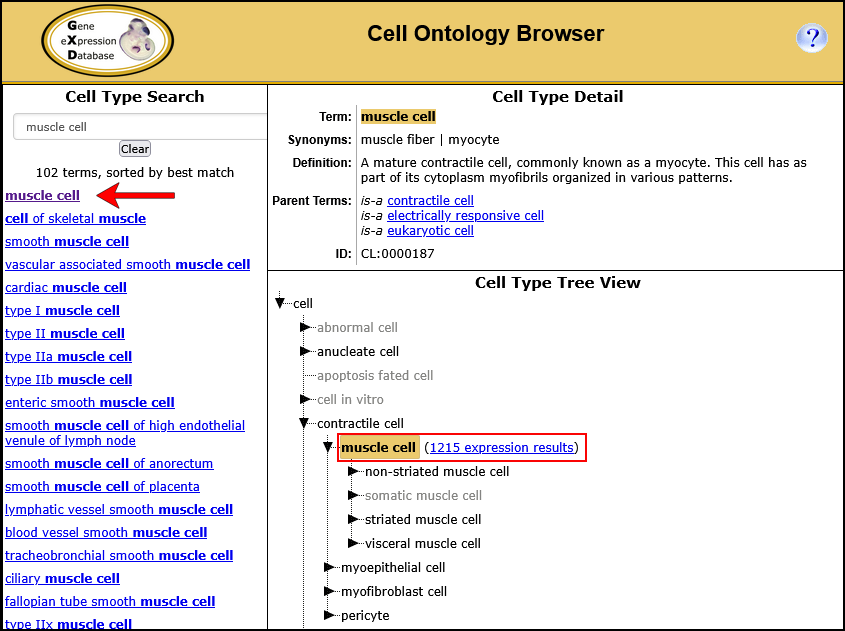
The Cell Ontology (CL) is a hierarchically structured, controlled vocabulary that describes a broad range of canonical biological cell types. You can use the Cell Ontology (CL) Browser to search for and view descriptions of cell types; to explore the ontology hierarchy and locate specific terms in context. You can then use the links provided to find the expression results associated with those terms.
|
||
| Accessing the Cell Ontology Browser | ||
|
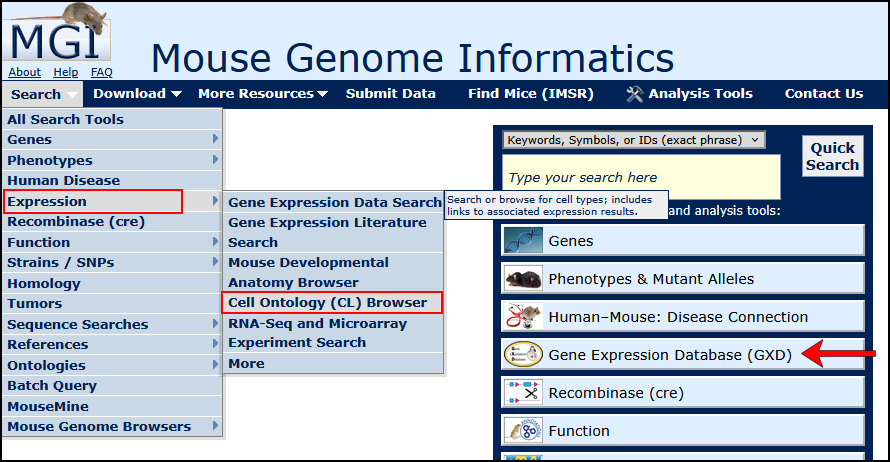
On most MGI pages, you can access the Cell Ontology Browser from the Search menu as shown in the image at right. Alternatively, you can also navigate to the Gene Expression Database (GXD) home page using the tile found on the MGI home page (arrow in image at right). Once there, click on the Cell Ontology Browser tile. For this tutorial, open the Cell Ontology Browser in a new window. Scroll down this page for further instructions. |
|
|
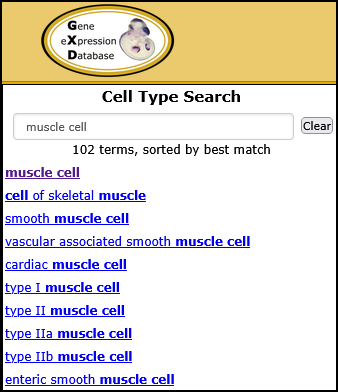
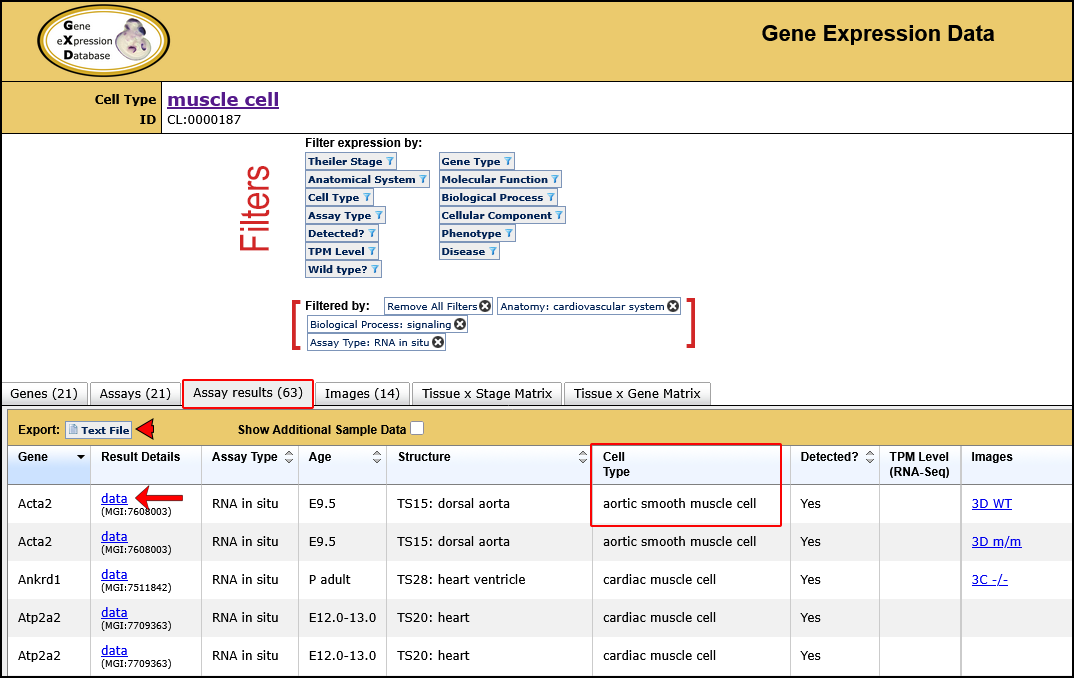
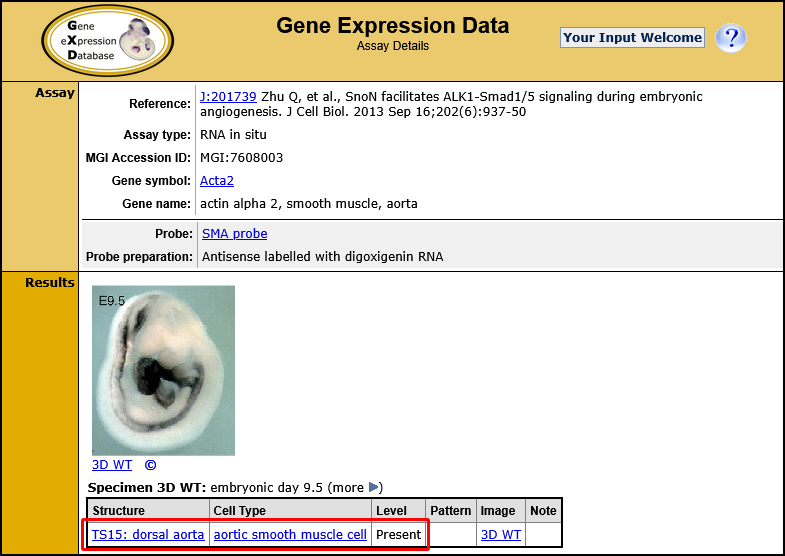
| Example. Find all gene expression results annotated to muscle cell and child terms. | ||
|
|
|
|
|
|
|
|
|
|
|
|
|
| |