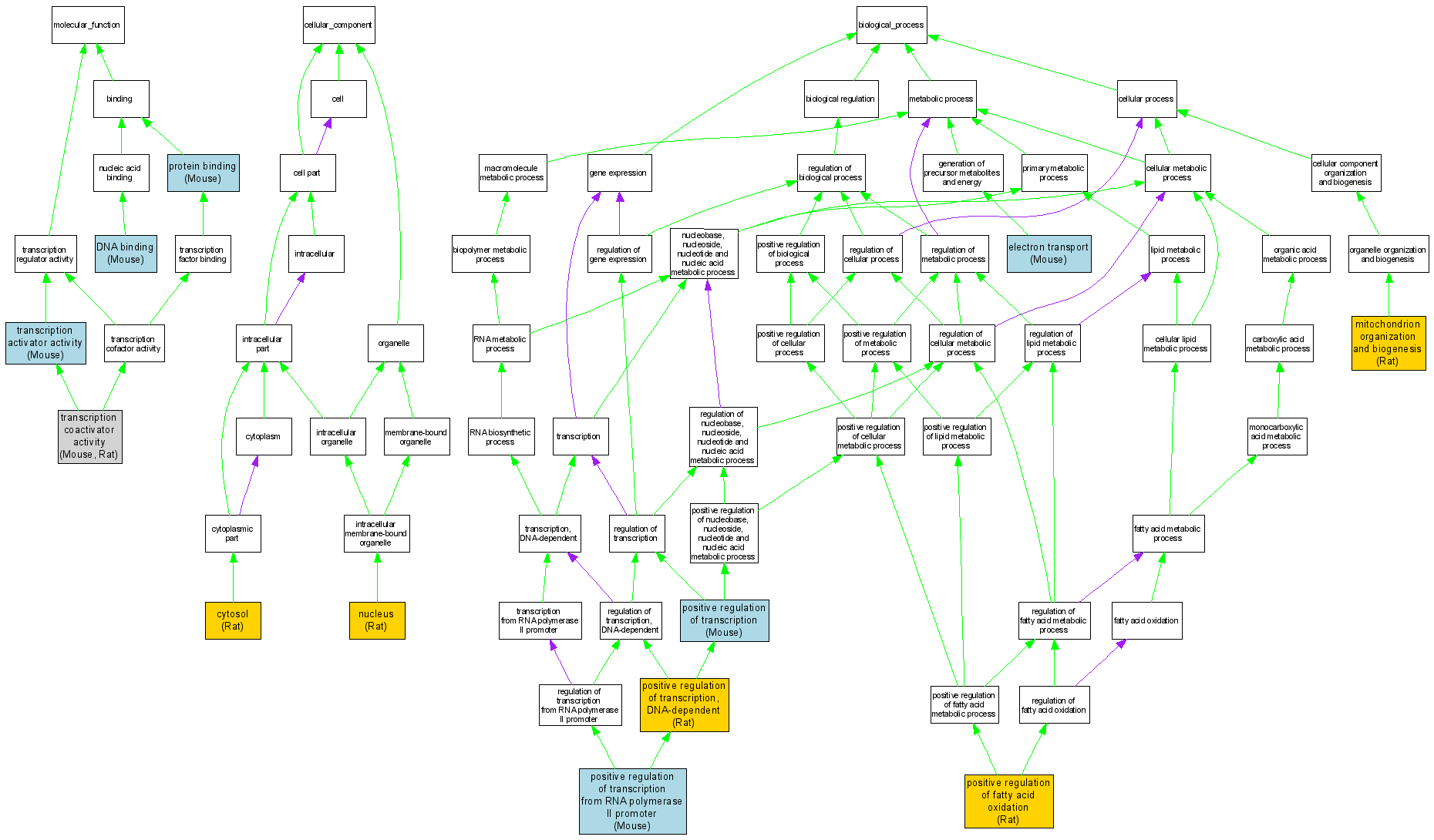
| Category | ID | Term | DB Object | Evidence | Organism | Reference |
| Molecular Function | GO:0003677 | DNA binding |
MGI:1342774 | IDA | Mouse | MGI:MGI:3529655|PMID:15681609 |
| Molecular Function | GO:0005515 | protein binding |
MGI:1342774 | IPI | Mouse | MGI:MGI:3689757|PMID:17050673 |
| Molecular Function | GO:0016563 | transcription activator activity |
MGI:1342774 | IDA | Mouse | MGI:MGI:3529655|PMID:15681609 |
| Molecular Function | GO:0003713 | transcription coactivator activity |
MGI:1342774 | IMP | Mouse | MGI:MGI:3689805|PMID:16271724 |
| Molecular Function | GO:0003713 | transcription coactivator activity |
RGD:620925 | IDA | Rat | RGD:633650|PMID:12107181 |
| Cellular Component | GO:0005829 | cytosol |
RGD:620925 | IDA | Rat | RGD:1601465|PMID:17099248 |
| Cellular Component | GO:0005634 | nucleus |
RGD:620925 | IDA | Rat | RGD:1601465|PMID:17099248 |
| Biological Process | GO:0006118 | electron transport |
MGI:1342774 | IDA | Mouse | MGI:MGI:3032469|PMID:14744933 |
| Biological Process | GO:0007005 | mitochondrion organization and biogenesis |
RGD:620925 | IMP | Rat | RGD:1601465|PMID:17099248 |
| Biological Process | GO:0046321 | positive regulation of fatty acid oxidation |
RGD:620925 | IMP | Rat | RGD:1601466|PMID:16139565 |
| Biological Process | GO:0045941 | positive regulation of transcription |
MGI:1342774 | IDA | Mouse | MGI:MGI:3032469|PMID:14744933 |
| Biological Process | GO:0045941 | positive regulation of transcription |
MGI:1342774 | IDA | Mouse | MGI:MGI:3529655|PMID:15681609 |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter |
MGI:1342774 | IGI | Mouse | MGI:MGI:3053497|PMID:15199055 |
| Biological Process | GO:0045893 | positive regulation of transcription, DNA-dependent |
RGD:620925 | IMP | Rat | RGD:1601466|PMID:16139565 |