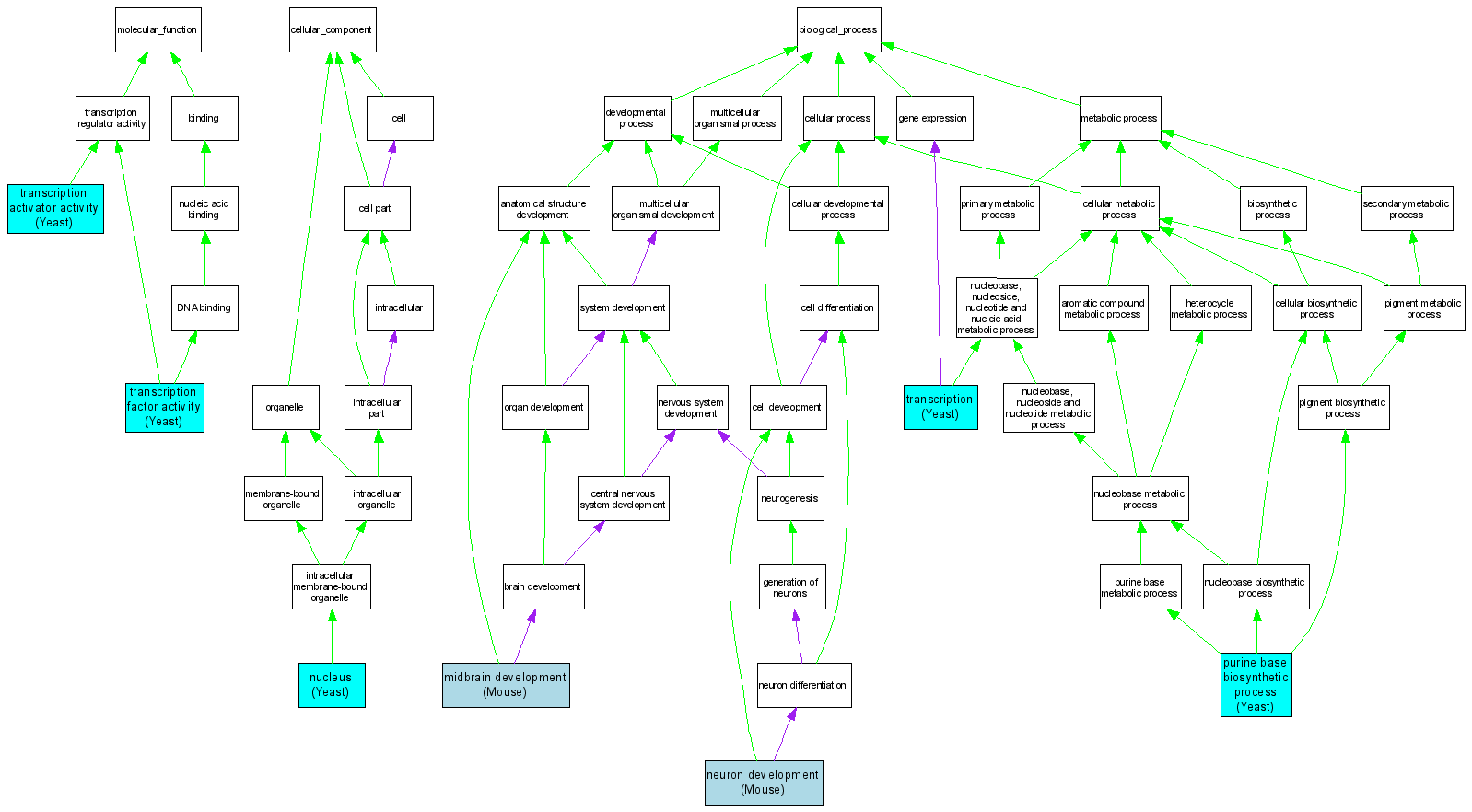
| Category | ID | Term | DB Object | Evidence | Organism | Reference |
| Molecular Function | GO:0016563 | transcription activator activity |
S000002264 | IMP | Yeast | SGD_REF:S000049914|PMID:3049251 |
| Molecular Function | GO:0003700 | transcription factor activity |
S000002264 | IMP | Yeast | SGD_REF:S000049914|PMID:3049251 |
| Molecular Function | GO:0003700 | transcription factor activity |
S000002264 | IGI | Yeast | SGD_REF:S000049914|PMID:3049251 |
| Molecular Function | GO:0003700 | transcription factor activity |
S000002264 | IPI | Yeast | SGD_REF:S000039230|PMID:7902583 |
| Cellular Component | GO:0005634 | nucleus |
S000002264 | IDA | Yeast | SGD_REF:S000047624|PMID:1493793 |
| Biological Process | GO:0030901 | midbrain development |
MGI:1100498 | IMP | Mouse | MGI:MGI:3035846|PMID:14973278 |
| Biological Process | GO:0048666 | neuron development |
MGI:1100498 | IMP | Mouse | MGI:MGI:3035846|PMID:14973278 |
| Biological Process | GO:0009113 | purine base biosynthetic process |
S000002264 | IDA | Yeast | SGD_REF:S000052116|PMID:1495962 |
| Biological Process | GO:0006350 | transcription |
S000002264 | IGI | Yeast | SGD_REF:S000071684|PMID:12145299 |