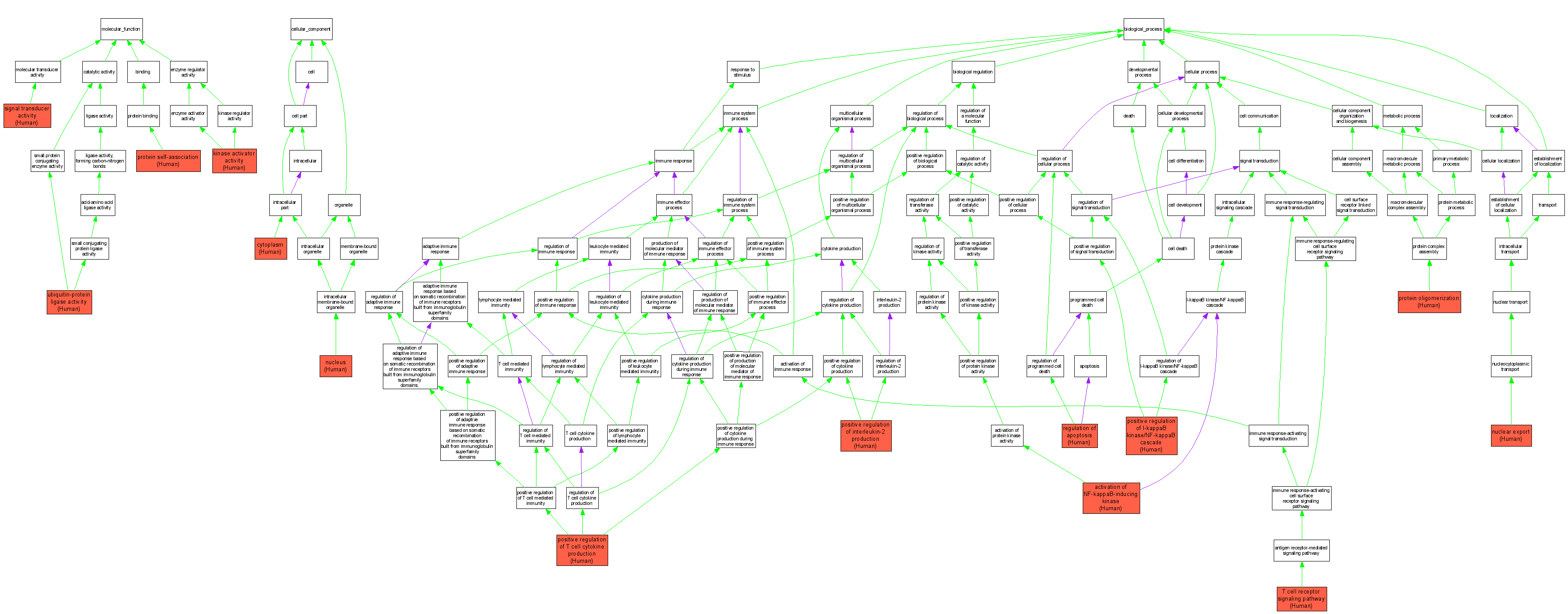
| Category | ID | Term | DB Object | Evidence | Organism | Reference |
| Molecular Function | GO:0019209 | kinase activator activity |
Q9UDY8 | IMP | Human | PMID:15125833 |
| Molecular Function | GO:0043621 | protein self-association |
Q9UDY8 | IPI | Human | PMID:15125833 |
| Molecular Function | GO:0004871 | signal transducer activity |
Q9UDY8 | IMP | Human | PMID:12761501 |
| Molecular Function | GO:0004842 | ubiquitin-protein ligase activity |
Q9UDY8 | IDA | Human | PMID:14695475 |
| Molecular Function | GO:0004842 | ubiquitin-protein ligase activity |
Q9UDY8 | IDA | Human | PMID:15125833 |
| Cellular Component | GO:0005737 | cytoplasm |
Q9UDY8 | IDA | Human | PMID:16123224 |
| Cellular Component | GO:0005634 | nucleus |
Q9UDY8 | IDA | Human | PMID:16123224 |
| Biological Process | GO:0007250 | activation of NF-kappaB-inducing kinase |
Q9UDY8 | IMP | Human | PMID:15125833 |
| Biological Process | GO:0051168 | nuclear export |
Q9UDY8 | IDA | Human | PMID:16123224 |
| Biological Process | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade |
Q9UDY8 | IMP | Human | PMID:12761501 |
| Biological Process | GO:0032743 | positive regulation of interleukin-2 production |
Q9UDY8 | IMP | Human | PMID:15125833 |
| Biological Process | GO:0002726 | positive regulation of T cell cytokine production |
Q9UDY8 | IMP | Human | PMID:15125833 |
| Biological Process | GO:0051259 | protein oligomerization |
Q9UDY8 | IDA | Human | PMID:15125833 |
| Biological Process | GO:0042981 | regulation of apoptosis |
Q9UDY8 | IDA | Human | PMID:12819136 |
| Biological Process | GO:0050852 | T cell receptor signaling pathway |
Q9UDY8 | IDA | Human | PMID:15125833 |