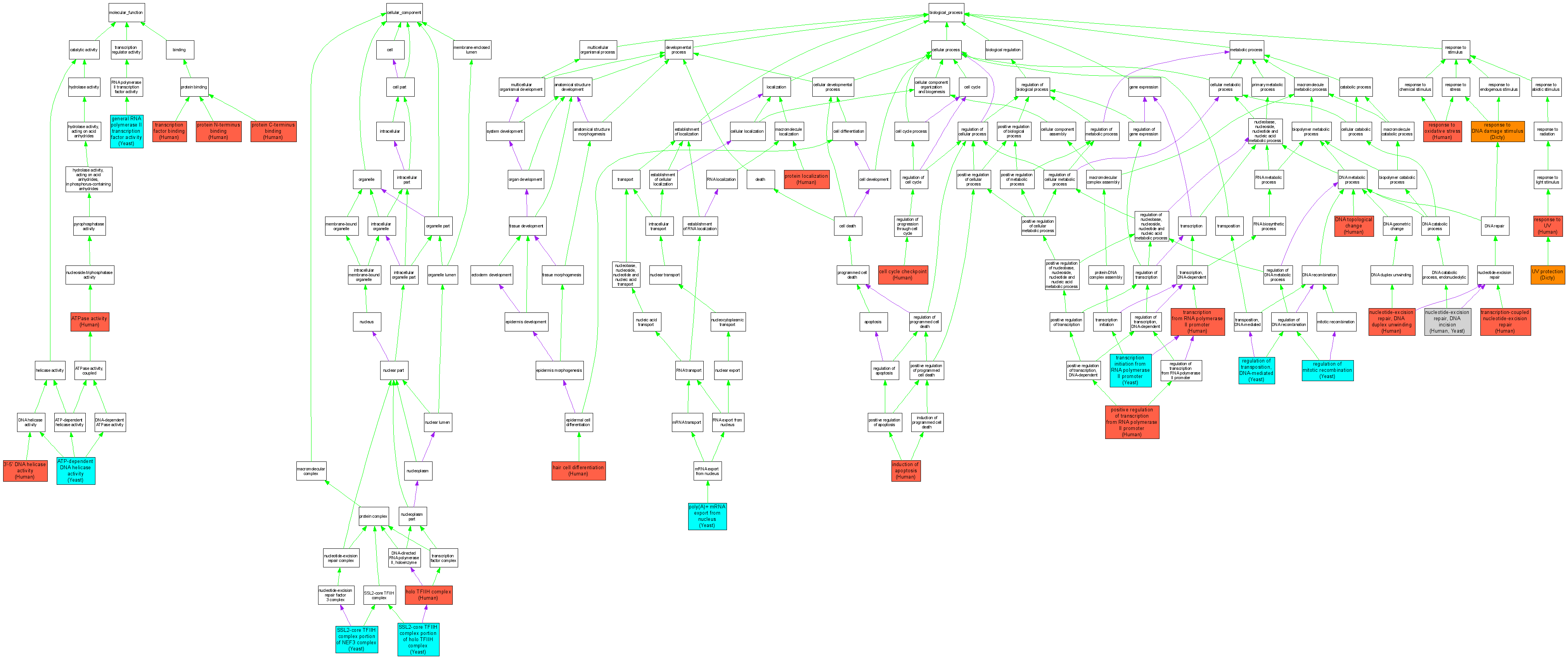
| Category | ID | Term | DB Object | Evidence | Organism | Reference |
| Molecular Function | GO:0043138 | 3'-5' DNA helicase activity |
P19447 | IMP | Human | PMID:8663148 |
| Molecular Function | GO:0043138 | 3'-5' DNA helicase activity |
P19447 | IDA | Human | PMID:17466626 |
| Molecular Function | GO:0004003 | ATP-dependent DNA helicase activity |
S000001405 | IDA | Yeast | SGD_REF:S000041779|PMID:8202161 |
| Molecular Function | GO:0016887 | ATPase activity |
P19447 | IDA | Human | PMID:17466626 |
| Molecular Function | GO:0016251 | general RNA polymerase II transcription factor activity |
S000001405 | IDA | Yeast | SGD_REF:S000053653|PMID:7813015 |
| Molecular Function | GO:0008022 | protein C-terminus binding |
P19447 | IPI | Human | PMID:10801852 |
| Molecular Function | GO:0008022 | protein C-terminus binding |
P19447 | IPI | Human | PMID:17466626 |
| Molecular Function | GO:0047485 | protein N-terminus binding |
P19447 | IPI | Human | PMID:8652557 |
| Molecular Function | GO:0008134 | transcription factor binding |
P19447 | IDA | Human | PMID:9173976 |
| Cellular Component | GO:0005675 | holo TFIIH complex |
P19447 | IDA | Human | PMID:8692842 |
| Cellular Component | GO:0000443 | SSL2-core TFIIH complex portion of holo TFIIH complex |
S000001405 | IDA | Yeast | SGD_REF:S000075290|PMID:14500720 |
| Cellular Component | GO:0000442 | SSL2-core TFIIH complex portion of NEF3 complex |
S000001405 | IDA | Yeast | SGD_REF:S000063827|PMID:8631896 |
| Biological Process | GO:0000075 | cell cycle checkpoint |
P19447 | IMP | Human | PMID:17088560 |
| Biological Process | GO:0006265 | DNA topological change |
P19447 | IMP | Human | PMID:8663148 |
| Biological Process | GO:0035315 | hair cell differentiation |
P19447 | IMP | Human | PMID:11335038 |
| Biological Process | GO:0006917 | induction of apoptosis |
P19447 | IMP | Human | PMID:8675009 |
| Biological Process | GO:0006917 | induction of apoptosis |
P19447 | IDA | Human | PMID:16914395 |
| Biological Process | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding |
P19447 | IMP | Human | PMID:17466626 |
| Biological Process | GO:0033683 | nucleotide-excision repair, DNA incision |
P19447 | IMP | Human | PMID:17466626 |
| Biological Process | GO:0033683 | nucleotide-excision repair, DNA incision |
P19447 | IMP | Human | PMID:8692841 |
| Biological Process | GO:0033683 | nucleotide-excision repair, DNA incision |
S000001405 | IDA | Yeast | SGD_REF:S000063827|PMID:8631896 |
| Biological Process | GO:0016973 | poly(A)+ mRNA export from nucleus |
S000001405 | IMP | Yeast | SGD_REF:S000120332|PMID:17212653 |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter |
P19447 | IDA | Human | PMID:8692841 |
| Biological Process | GO:0008104 | protein localization |
P19447 | IMP | Human | PMID:17509950 |
| Biological Process | GO:0000019 | regulation of mitotic recombination |
S000001405 | IMP | Yeast | SGD_REF:S000051731|PMID:10713167 |
| Biological Process | GO:0000337 | regulation of transposition, DNA-mediated |
S000001405 | IMP | Yeast | SGD_REF:S000051731|PMID:10713167 |
| Biological Process | GO:0006974 | response to DNA damage stimulus |
DDB0214830 | IEP | Dicty | DDB_REF:7171|PMID:9649625 |
| Biological Process | GO:0006979 | response to oxidative stress |
P19447 | IMP | Human | PMID:17614221 |
| Biological Process | GO:0009411 | response to UV |
P19447 | IMP | Human | PMID:17509950 |
| Biological Process | GO:0006366 | transcription from RNA polymerase II promoter |
P19447 | IMP | Human | PMID:8663148 |
| Biological Process | GO:0006367 | transcription initiation from RNA polymerase II promoter |
S000001405 | IMP | Yeast | SGD_REF:S000045282|PMID:10409754 |
| Biological Process | GO:0006367 | transcription initiation from RNA polymerase II promoter |
S000001405 | IMP | Yeast | SGD_REF:S000053530|PMID:10713451 |
| Biological Process | GO:0006283 | transcription-coupled nucleotide-excision repair |
P19447 | IDA | Human | PMID:8663148 |
| Biological Process | GO:0009650 | UV protection |
DDB0214830 | IMP | Dicty | DDB_REF:3606|PMID:9765592 |