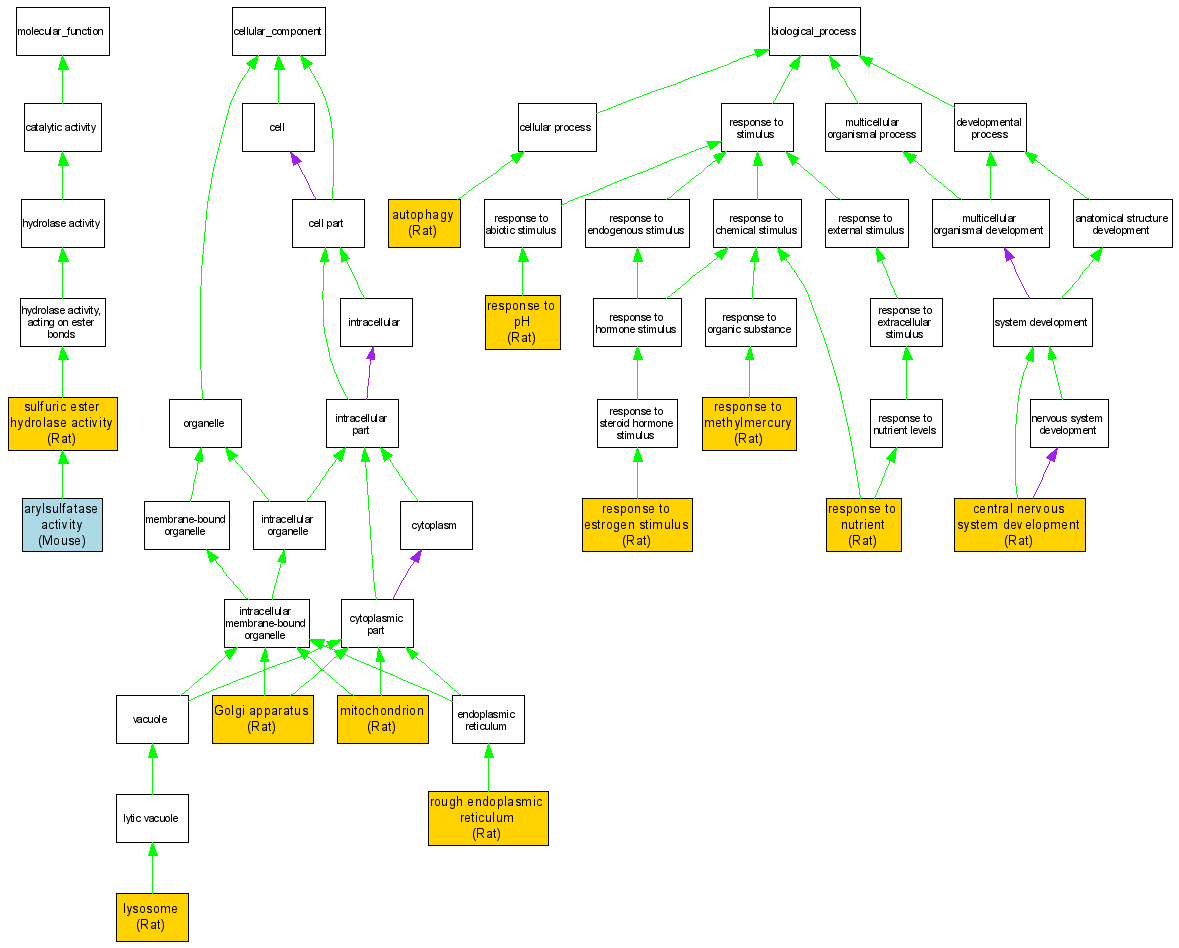
| Category | ID | Term | DB Object | Evidence | Organism | Reference |
| Molecular Function | GO:0004065 | arylsulfatase activity |
MGI:88075 | IDA | Mouse | MGI:MGI:54939|PMID:7470017 |
| Molecular Function | GO:0008484 | sulfuric ester hydrolase activity |
RGD:2158 | IDA | Rat | RGD:1599231|PMID:2884099 |
| Cellular Component | GO:0005794 | Golgi apparatus |
RGD:2158 | IDA | Rat | RGD:1599233|PMID:7419898 |
| Cellular Component | GO:0005764 | lysosome |
RGD:2158 | IDA | Rat | RGD:1599229|PMID:1686699 |
| Cellular Component | GO:0005739 | mitochondrion |
RGD:2158 | IDA | Rat | RGD:1599237|PMID:5544743 |
| Cellular Component | GO:0005791 | rough endoplasmic reticulum |
RGD:2158 | IDA | Rat | RGD:1599233|PMID:7419898 |
| Biological Process | GO:0006914 | autophagy |
RGD:2158 | IDA | Rat | RGD:1599235|PMID:8037 |
| Biological Process | GO:0007417 | central nervous system development |
RGD:2158 | IDA | Rat | RGD:1599226|PMID:1671824 |
| Biological Process | GO:0043627 | response to estrogen stimulus |
RGD:2158 | IDA | Rat | RGD:1599234|PMID:15040 |
| Biological Process | GO:0051597 | response to methylmercury |
RGD:2158 | IDA | Rat | RGD:1599225|PMID:1682910 |
| Biological Process | GO:0007584 | response to nutrient |
RGD:2158 | IDA | Rat | RGD:1599230|PMID:2290353 |
| Biological Process | GO:0009268 | response to pH |
RGD:2158 | IDA | Rat | RGD:1599227|PMID:6137211 |