|
Symbol Name ID |
Ighmbp2
immunoglobulin mu DNA binding protein 2 MGI:99954 |
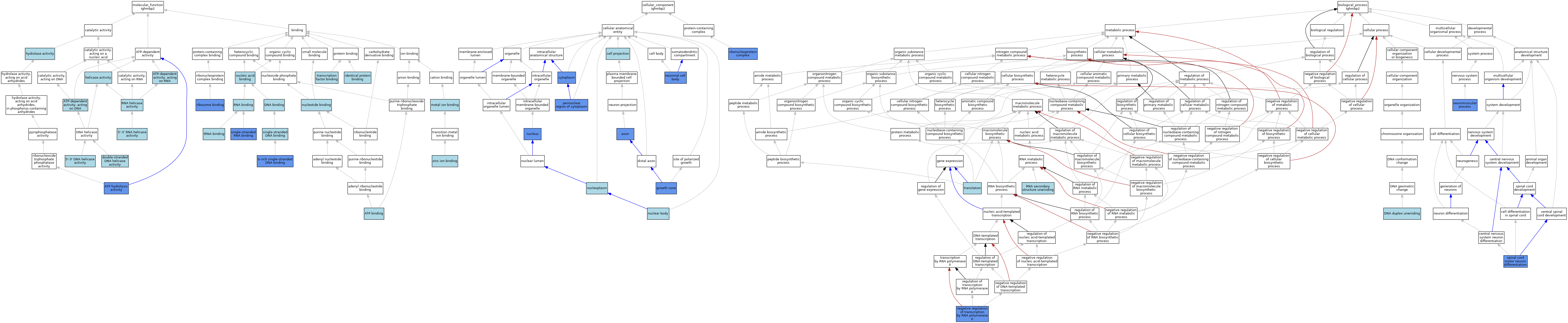
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0043139 | 5'-3' DNA helicase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043139 | 5'-3' DNA helicase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0032574 | 5'-3' RNA helicase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IDA | J:314557 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008094 | ATP-dependent activity, acting on DNA | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008186 | ATP-dependent activity, acting on RNA | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0036121 | double-stranded DNA helicase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990955 | G-rich single-stranded DNA binding | IDA | J:4973 | |||||||||
| Molecular Function | GO:0004386 | helicase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004386 | helicase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:148546 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0043022 | ribosome binding | IDA | J:146154 | |||||||||
| Molecular Function | GO:0043022 | ribosome binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003724 | RNA helicase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003727 | single-stranded RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003727 | single-stranded RNA binding | IDA | J:314557 | |||||||||
| Molecular Function | GO:0008134 | transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000049 | tRNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000049 | tRNA binding | ISO | J:148546 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:146154 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:148546 | |||||||||
| Cellular Component | GO:0030426 | growth cone | IDA | J:146154 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:92862 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:92862 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:148546 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IDA | J:146154 | |||||||||
| Cellular Component | GO:1990904 | ribonucleoprotein complex | IDA | J:146154 | |||||||||
| Biological Process | GO:0032508 | DNA duplex unwinding | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:155856 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:91301 | |||||||||
| Biological Process | GO:0050905 | neuromuscular process | IMP | J:99795 | |||||||||
| Biological Process | GO:0010501 | RNA secondary structure unwinding | ISO | J:164563 | |||||||||
| Biological Process | GO:0021522 | spinal cord motor neuron differentiation | IMP | J:148546 | |||||||||
| Biological Process | GO:0006412 | translation | NAS | J:148546 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||