|
Symbol Name ID |
Hsp90b1
heat shock protein 90, beta (Grp94), member 1 MGI:98817 |
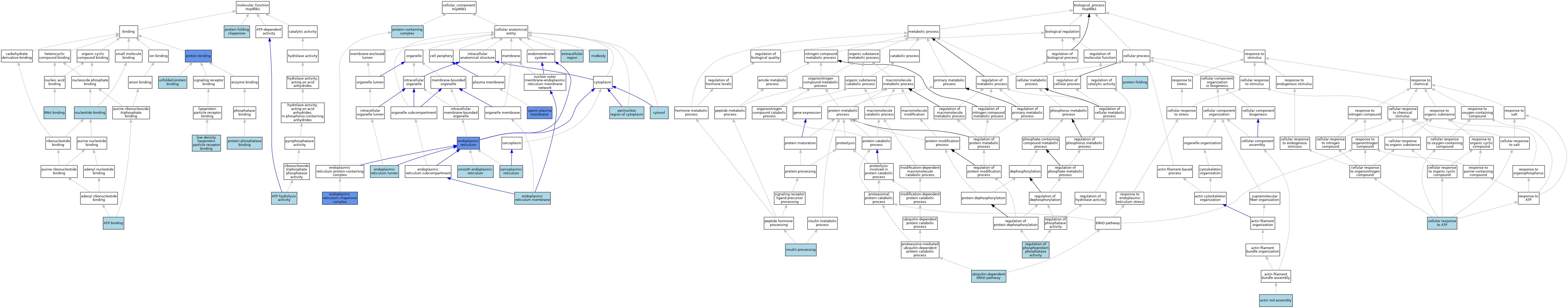
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0050750 | low-density lipoprotein particle receptor binding | ISO | J:90197 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:167004 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:193640 | |||||||||
| Molecular Function | GO:0044183 | protein folding chaperone | ISO | J:327398 | |||||||||
| Molecular Function | GO:0019903 | protein phosphatase binding | ISO | J:142802 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051082 | unfolded protein binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:268046 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:78076 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:92989 | |||||||||
| Cellular Component | GO:0034663 | endoplasmic reticulum chaperone complex | IDA | J:217288 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | ISO | J:327398 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-2173779 | |||||||||
| Cellular Component | GO:0030496 | midbody | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0097524 | sperm plasma membrane | IDA | J:91263 | |||||||||
| Biological Process | GO:0031247 | actin rod assembly | ISO | J:142802 | |||||||||
| Biological Process | GO:0071318 | cellular response to ATP | ISO | J:142802 | |||||||||
| Biological Process | GO:0030070 | insulin processing | ISO | J:327398 | |||||||||
| Biological Process | GO:0006457 | protein folding | IBA | J:265628 | |||||||||
| Biological Process | GO:0043666 | regulation of phosphoprotein phosphatase activity | ISO | J:142802 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||