|
Symbol Name ID |
Thrb
thyroid hormone receptor beta MGI:98743 |
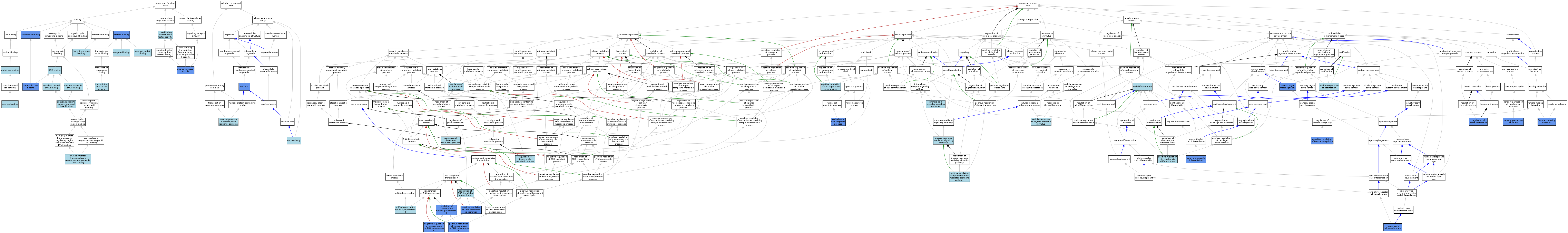
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:178620 | |||||||||
| Molecular Function | GO:0031490 | chromatin DNA binding | IDA | J:130306 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | TAS | J:52829 | |||||||||
| Molecular Function | GO:0003690 | double-stranded DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | IMP | J:159022 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | TAS | J:42911 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | TAS | J:63101 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | TAS | J:52829 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | TAS | J:51466 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | TAS | J:56668 | |||||||||
| Molecular Function | GO:0004879 | nuclear receptor activity | IMP | J:159022 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:115974 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:218799 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:142045 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:46724 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:125047 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070324 | thyroid hormone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001223 | transcription coactivator binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | TAS | J:51466 | |||||||||
| Cellular Component | GO:0005634 | nucleus | TAS | J:42911 | |||||||||
| Cellular Component | GO:0005634 | nucleus | TAS | J:52829 | |||||||||
| Cellular Component | GO:0005634 | nucleus | TAS | J:56668 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:130306 | |||||||||
| Cellular Component | GO:0090575 | RNA polymerase II transcription regulator complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0009887 | animal organ morphogenesis | IMP | J:52829 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0097067 | cellular response to thyroid hormone stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0008050 | female courtship behavior | IMP | J:61839 | |||||||||
| Biological Process | GO:0042789 | mRNA transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IDA | J:87883 | |||||||||
| Biological Process | GO:0007621 | negative regulation of female receptivity | IMP | J:61839 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:29044 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:178114 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0032332 | positive regulation of chondrocyte differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0045778 | positive regulation of ossification | ISO | J:155856 | |||||||||
| Biological Process | GO:0002157 | positive regulation of thyroid hormone mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:108438 | |||||||||
| Biological Process | GO:0090181 | regulation of cholesterol metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | TAS | J:52829 | |||||||||
| Biological Process | GO:0008016 | regulation of heart contraction | IMP | J:51466 | |||||||||
| Biological Process | GO:0019216 | regulation of lipid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IGI | J:142045 | |||||||||
| Biological Process | GO:0090207 | regulation of triglyceride metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0097474 | retinal cone cell apoptotic process | IMP | J:159022 | |||||||||
| Biological Process | GO:0046549 | retinal cone cell development | IMP | J:159022 | |||||||||
| Biological Process | GO:0048384 | retinoic acid receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IMP | J:63101 | |||||||||
| Biological Process | GO:0002154 | thyroid hormone mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0060509 | type I pneumocyte differentiation | IGI | J:178114 | |||||||||
| Biological Process | GO:0060509 | type I pneumocyte differentiation | IGI | J:178114 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||