|
Symbol Name ID |
Srsf1
serine and arginine-rich splicing factor 1 MGI:98283 |
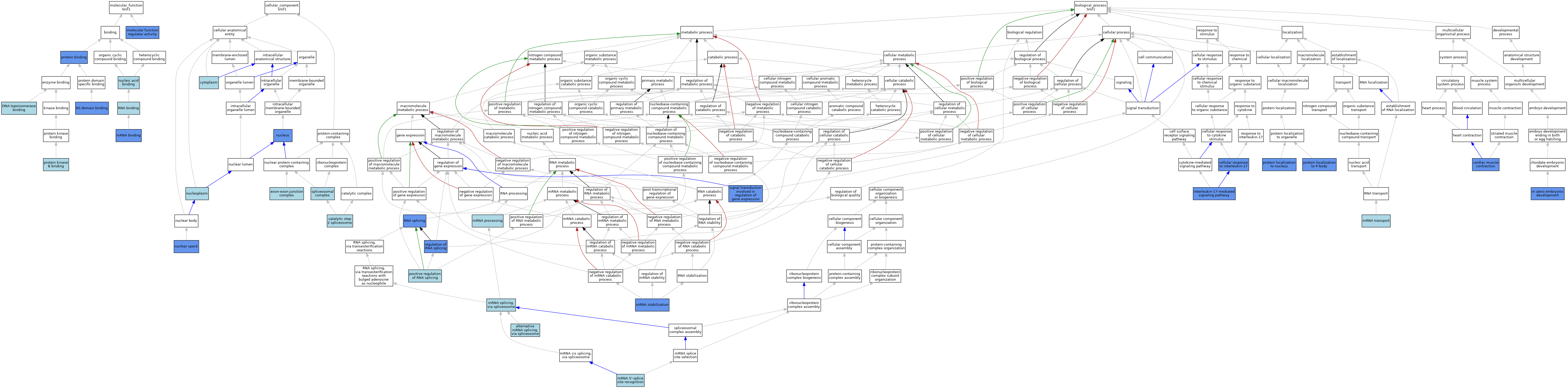
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0044547 | DNA topoisomerase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0098772 | molecular function regulator activity | IDA | J:176465 | |||||||||
| Molecular Function | GO:0003729 | mRNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003729 | mRNA binding | IMP | J:176465 | |||||||||
| Molecular Function | GO:0003729 | mRNA binding | ISO | J:197742 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:206891 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200368 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:314085 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:188557 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:282433 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:176465 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:176465 | |||||||||
| Molecular Function | GO:0043422 | protein kinase B binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0050733 | RS domain binding | IPI | J:97604 | |||||||||
| Cellular Component | GO:0071013 | catalytic step 2 spliceosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0035145 | exon-exon junction complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | IDA | J:171647 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | IDA | J:164388 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:314085 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IPI | J:282433 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:197742 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:94966 | |||||||||
| Cellular Component | GO:0005681 | spliceosomal complex | IEA | J:60000 | |||||||||
| Biological Process | GO:0000380 | alternative mRNA splicing, via spliceosome | ISO | J:155856 | |||||||||
| Biological Process | GO:0000380 | alternative mRNA splicing, via spliceosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0060048 | cardiac muscle contraction | IGI | J:96039 | |||||||||
| Biological Process | GO:0060048 | cardiac muscle contraction | IGI | J:96039 | |||||||||
| Biological Process | GO:0097398 | cellular response to interleukin-17 | IDA | J:282433 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:96039 | |||||||||
| Biological Process | GO:0097400 | interleukin-17-mediated signaling pathway | IMP | J:176465 | |||||||||
| Biological Process | GO:0097400 | interleukin-17-mediated signaling pathway | IDA | J:176465 | |||||||||
| Biological Process | GO:0000395 | mRNA 5'-splice site recognition | ISO | J:164563 | |||||||||
| Biological Process | GO:0006397 | mRNA processing | IEA | J:60000 | |||||||||
| Biological Process | GO:0000398 | mRNA splicing, via spliceosome | IBA | J:265628 | |||||||||
| Biological Process | GO:0048255 | mRNA stabilization | IMP | J:176465 | |||||||||
| Biological Process | GO:0048255 | mRNA stabilization | IDA | J:176465 | |||||||||
| Biological Process | GO:0051028 | mRNA transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0033120 | positive regulation of RNA splicing | ISO | J:155856 | |||||||||
| Biological Process | GO:0034504 | protein localization to nucleus | IDA | J:282433 | |||||||||
| Biological Process | GO:0110012 | protein localization to P-body | IDA | J:282433 | |||||||||
| Biological Process | GO:0043484 | regulation of RNA splicing | IMP | J:314085 | |||||||||
| Biological Process | GO:0008380 | RNA splicing | IGI | J:96039 | |||||||||
| Biological Process | GO:0008380 | RNA splicing | IDA | J:97604 | |||||||||
| Biological Process | GO:0008380 | RNA splicing | IGI | J:96039 | |||||||||
| Biological Process | GO:0023019 | signal transduction involved in regulation of gene expression | IDA | J:176465 | |||||||||
| Biological Process | GO:0023019 | signal transduction involved in regulation of gene expression | IMP | J:176465 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||