|
Symbol Name ID |
Ccl3
C-C motif chemokine ligand 3 MGI:98260 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
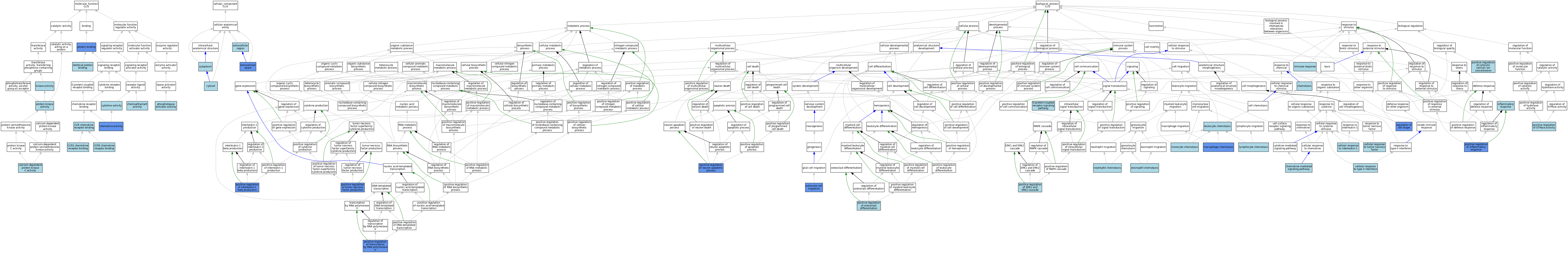
| Molecular Function | GO:0004698 | calcium-dependent protein kinase C activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0048020 | CCR chemokine receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031726 | CCR1 chemokine receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031730 | CCR5 chemokine receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042056 | chemoattractant activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008009 | chemokine activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008009 | chemokine activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008009 | chemokine activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008009 | chemokine activity | IDA | J:29873 | |||||||||
| Molecular Function | GO:0005125 | cytokine activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016004 | phospholipase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:247428 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:29873 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:40322 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:29873 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:194920 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:232381 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:147177 | |||||||||
| Biological Process | GO:0043615 | astrocyte cell migration | IDA | J:90687 | |||||||||
| Biological Process | GO:0071347 | cellular response to interleukin-1 | IBA | J:265628 | |||||||||
| Biological Process | GO:0071356 | cellular response to tumor necrosis factor | IBA | J:265628 | |||||||||
| Biological Process | GO:0071346 | cellular response to type II interferon | IBA | J:265628 | |||||||||
| Biological Process | GO:0070098 | chemokine-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0006935 | chemotaxis | IEA | J:60000 | |||||||||
| Biological Process | GO:0048245 | eosinophil chemotaxis | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0006955 | immune response | IEA | J:72247 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IBA | J:265628 | |||||||||
| Biological Process | GO:0030595 | leukocyte chemotaxis | ISO | J:155856 | |||||||||
| Biological Process | GO:0048247 | lymphocyte chemotaxis | IBA | J:265628 | |||||||||
| Biological Process | GO:0048246 | macrophage chemotaxis | IGI | J:179147 | |||||||||
| Biological Process | GO:0002548 | monocyte chemotaxis | IBA | J:265628 | |||||||||
| Biological Process | GO:0030593 | neutrophil chemotaxis | ISO | J:155856 | |||||||||
| Biological Process | GO:0030593 | neutrophil chemotaxis | IBA | J:265628 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | IBA | J:265628 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0050729 | positive regulation of inflammatory response | IGI | J:179147 | |||||||||
| Biological Process | GO:0032731 | positive regulation of interleukin-1 beta production | IGI | J:179147 | |||||||||
| Biological Process | GO:0043525 | positive regulation of neuron apoptotic process | IGI | J:90687 | |||||||||
| Biological Process | GO:0045672 | positive regulation of osteoclast differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:90687 | |||||||||
| Biological Process | GO:0032760 | positive regulation of tumor necrosis factor production | IGI | J:179147 | |||||||||
| Biological Process | GO:0008360 | regulation of cell shape | IDA | J:90687 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||