|
Symbol Name ID |
Ros1
Ros1 proto-oncogene MGI:97999 |
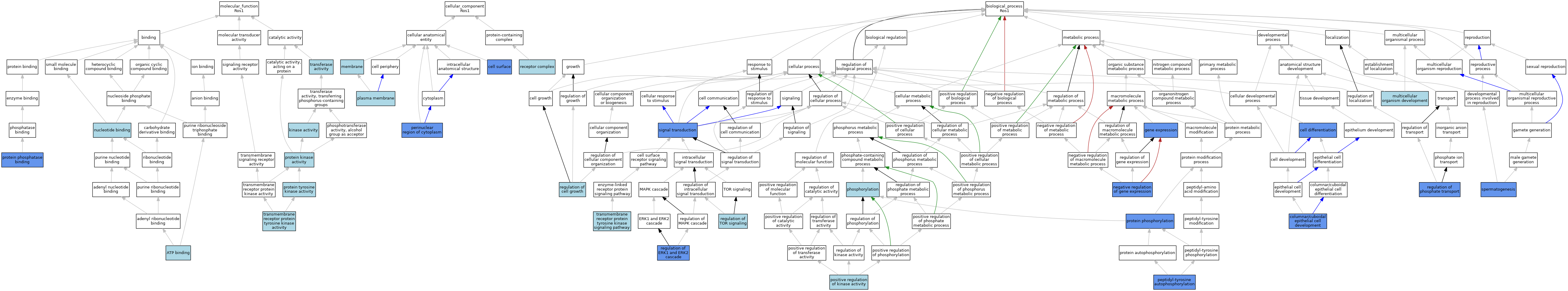
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0019903 | protein phosphatase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019903 | protein phosphatase binding | IPI | J:67582 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:21839 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IDA | J:21839 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IDA | J:67582 | |||||||||
| Biological Process | GO:0002066 | columnar/cuboidal epithelial cell development | IMP | J:33259 | |||||||||
| Biological Process | GO:0010467 | gene expression | IMP | J:106001 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IBA | J:265628 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | IMP | J:85614 | |||||||||
| Biological Process | GO:0038083 | peptidyl-tyrosine autophosphorylation | IDA | J:21839 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0033674 | positive regulation of kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:67582 | |||||||||
| Biological Process | GO:0001558 | regulation of cell growth | ISO | J:164563 | |||||||||
| Biological Process | GO:0070372 | regulation of ERK1 and ERK2 cascade | IDA | J:67582 | |||||||||
| Biological Process | GO:0010966 | regulation of phosphate transport | IMP | J:85614 | |||||||||
| Biological Process | GO:0032006 | regulation of TOR signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0032006 | regulation of TOR signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IDA | J:21389 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:33259 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:85614 | |||||||||
| Biological Process | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||