|
Symbol Name ID |
Pdha1
pyruvate dehydrogenase E1 alpha 1 MGI:97532 |
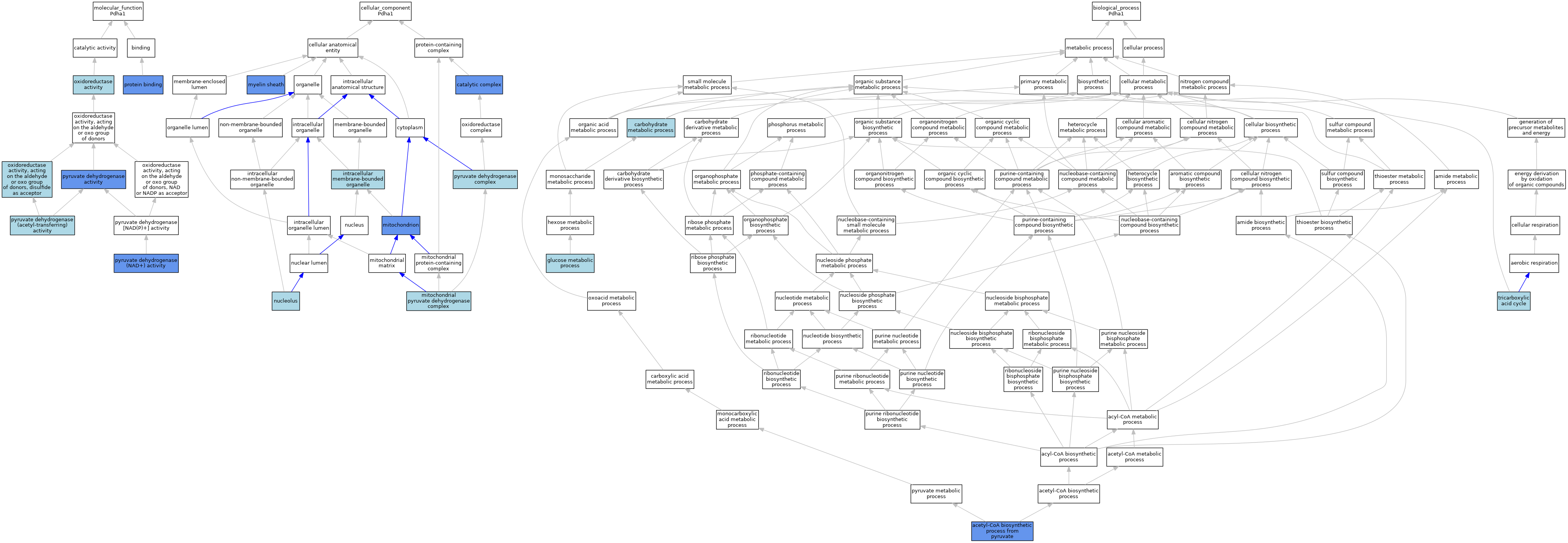
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:263469 | |||||||||
| Molecular Function | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0034604 | pyruvate dehydrogenase (NAD+) activity | IMP | J:141289 | |||||||||
| Molecular Function | GO:0034604 | pyruvate dehydrogenase (NAD+) activity | IMP | J:141289 | |||||||||
| Molecular Function | GO:0004738 | pyruvate dehydrogenase activity | IMP | J:74192 | |||||||||
| Cellular Component | GO:1902494 | catalytic complex | IMP | J:74192 | |||||||||
| Cellular Component | GO:1902494 | catalytic complex | IMP | J:123893 | |||||||||
| Cellular Component | GO:1902494 | catalytic complex | IMP | J:94805 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005967 | mitochondrial pyruvate dehydrogenase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:242350 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | HDA | J:145263 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045254 | pyruvate dehydrogenase complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045254 | pyruvate dehydrogenase complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | ISO | J:164563 | |||||||||
| Biological Process | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | IBA | J:265628 | |||||||||
| Biological Process | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | IMP | J:141289 | |||||||||
| Biological Process | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | IMP | J:141289 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006099 | tricarboxylic acid cycle | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||