|
Symbol Name ID |
Pcna
proliferating cell nuclear antigen MGI:97503 |
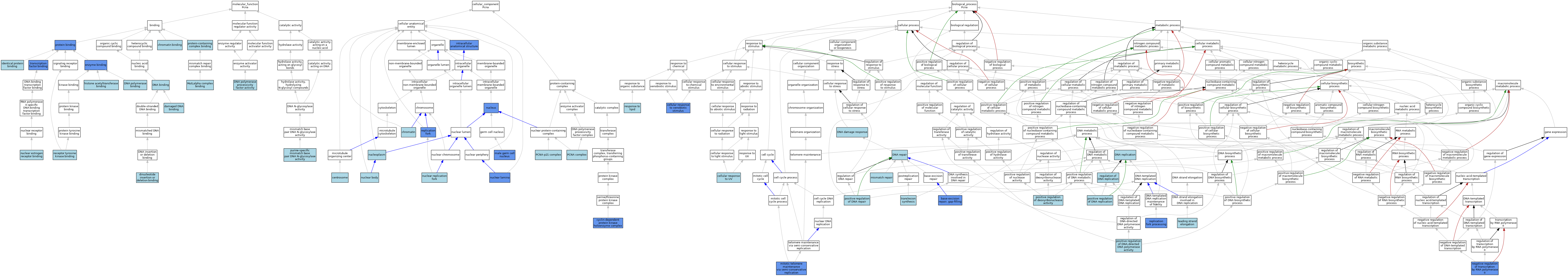
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0032139 | dinucleotide insertion or deletion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0070182 | DNA polymerase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030337 | DNA polymerase processivity factor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | IPI | J:202812 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035035 | histone acetyltransferase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0032405 | MutLalpha complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030331 | nuclear estrogen receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:121214 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:117431 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:248211 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:81304 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:78410 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:85809 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030971 | receptor tyrosine kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008134 | transcription factor binding | IPI | J:142927 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:204810 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IPI | J:85809 | |||||||||
| Cellular Component | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IPI | J:85809 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | IDA | J:106653 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:82276 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:82276 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005652 | nuclear lamina | IDA | J:81304 | |||||||||
| Cellular Component | GO:0043596 | nuclear replication fork | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:202812 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:162095 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:85809 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:105978 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:142136 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:76874 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:53290 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:88220 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:142927 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:204433 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:195630 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:142927 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:79935 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:78856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISS | J:93906 | |||||||||
| Cellular Component | GO:0043626 | PCNA complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043626 | PCNA complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0070557 | PCNA-p21 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005657 | replication fork | ISO | J:113839 | |||||||||
| Cellular Component | GO:0005657 | replication fork | IDA | J:76360 | |||||||||
| Biological Process | GO:0006287 | base-excision repair, gap-filling | IDA | J:91067 | |||||||||
| Biological Process | GO:0034644 | cellular response to UV | ISO | J:164563 | |||||||||
| Biological Process | GO:0071466 | cellular response to xenobiotic stimulus | IDA | J:210952 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0006260 | DNA replication | IEA | J:60000 | |||||||||
| Biological Process | GO:0006272 | leading strand elongation | IBA | J:265628 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | IBA | J:265628 | |||||||||
| Biological Process | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | IGI | J:202812 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:142927 | |||||||||
| Biological Process | GO:0032077 | positive regulation of deoxyribonuclease activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0045739 | positive regulation of DNA repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0045740 | positive regulation of DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0006275 | regulation of DNA replication | IEA | J:72247 | |||||||||
| Biological Process | GO:0031297 | replication fork processing | IGI | J:202812 | |||||||||
| Biological Process | GO:0033993 | response to lipid | ISO | J:155856 | |||||||||
| Biological Process | GO:0019985 | translesion synthesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0019985 | translesion synthesis | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||