|
Symbol Name ID |
Nf2
neurofibromin 2 MGI:97307 |
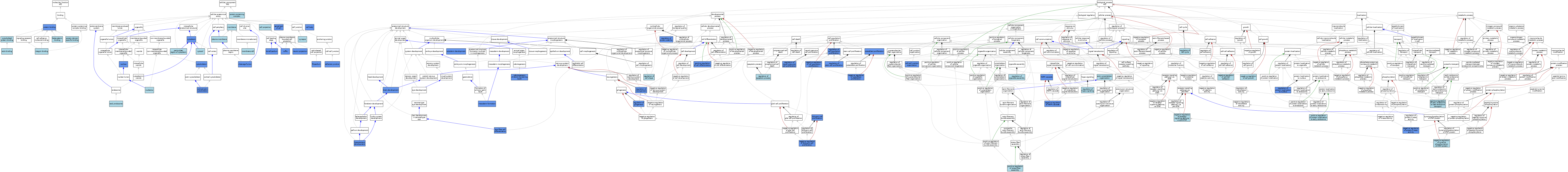
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008013 | beta-catenin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008092 | cytoskeletal protein binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:213963 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:213963 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:83082 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:86235 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104992 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IMP | J:83082 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:199301 | |||||||||
| Cellular Component | GO:0044297 | cell body | IDA | J:218609 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032154 | cleavage furrow | IDA | J:46198 | |||||||||
| Cellular Component | GO:0030864 | cortical actin cytoskeleton | IDA | J:94716 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:31660 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:31660 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:31660 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:91460 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IDA | J:94716 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030175 | filopodium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030175 | filopodium | IDA | J:46918 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:46918 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | IDA | J:218609 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:91460 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IDA | J:46918 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:199301 | |||||||||
| Biological Process | GO:0045216 | cell-cell junction organization | IMP | J:83082 | |||||||||
| Biological Process | GO:0007398 | ectoderm development | IMP | J:40596 | |||||||||
| Biological Process | GO:0021766 | hippocampus development | IMP | J:199301 | |||||||||
| Biological Process | GO:0070306 | lens fiber cell differentiation | IMP | J:161172 | |||||||||
| Biological Process | GO:0000165 | MAPK cascade | IMP | J:104992 | |||||||||
| Biological Process | GO:0001707 | mesoderm formation | IMP | J:40596 | |||||||||
| Biological Process | GO:0030308 | negative regulation of cell growth | ISO | J:155856 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IMP | J:47282 | |||||||||
| Biological Process | GO:0022408 | negative regulation of cell-cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0043409 | negative regulation of MAPK cascade | IMP | J:104992 | |||||||||
| Biological Process | GO:0033689 | negative regulation of osteoblast proliferation | IMP | J:63264 | |||||||||
| Biological Process | GO:0006469 | negative regulation of protein kinase activity | IDA | J:86235 | |||||||||
| Biological Process | GO:0006469 | negative regulation of protein kinase activity | IMP | J:104992 | |||||||||
| Biological Process | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | ISO | J:164563 | |||||||||
| Biological Process | GO:0010626 | negative regulation of Schwann cell proliferation | IMP | J:63264 | |||||||||
| Biological Process | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | ISO | J:164563 | |||||||||
| Biological Process | GO:0042475 | odontogenesis of dentin-containing tooth | IMP | J:63264 | |||||||||
| Biological Process | GO:0033687 | osteoblast proliferation | IMP | J:63264 | |||||||||
| Biological Process | GO:0045597 | positive regulation of cell differentiation | IMP | J:199301 | |||||||||
| Biological Process | GO:2000643 | positive regulation of early endosome to late endosome transport | IBA | J:265628 | |||||||||
| Biological Process | GO:1902966 | positive regulation of protein localization to early endosome | IBA | J:265628 | |||||||||
| Biological Process | GO:0051496 | positive regulation of stress fiber assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0042127 | regulation of cell population proliferation | IMP | J:83082 | |||||||||
| Biological Process | GO:0008360 | regulation of cell shape | IBA | J:265628 | |||||||||
| Biological Process | GO:0014013 | regulation of gliogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0014013 | regulation of gliogenesis | IMP | J:218609 | |||||||||
| Biological Process | GO:0035330 | regulation of hippo signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:2000177 | regulation of neural precursor cell proliferation | IMP | J:199301 | |||||||||
| Biological Process | GO:0050767 | regulation of neurogenesis | IMP | J:218609 | |||||||||
| Biological Process | GO:1902115 | regulation of organelle assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:1900180 | regulation of protein localization to nucleus | IMP | J:199301 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | IMP | J:199301 | |||||||||
| Biological Process | GO:0072091 | regulation of stem cell proliferation | IGI | J:218609 | |||||||||
| Biological Process | GO:0072091 | regulation of stem cell proliferation | IMP | J:218609 | |||||||||
| Biological Process | GO:0014010 | Schwann cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0014010 | Schwann cell proliferation | IMP | J:63264 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||