|
Symbol Name ID |
Mertk
MER proto-oncogene tyrosine kinase MGI:96965 |
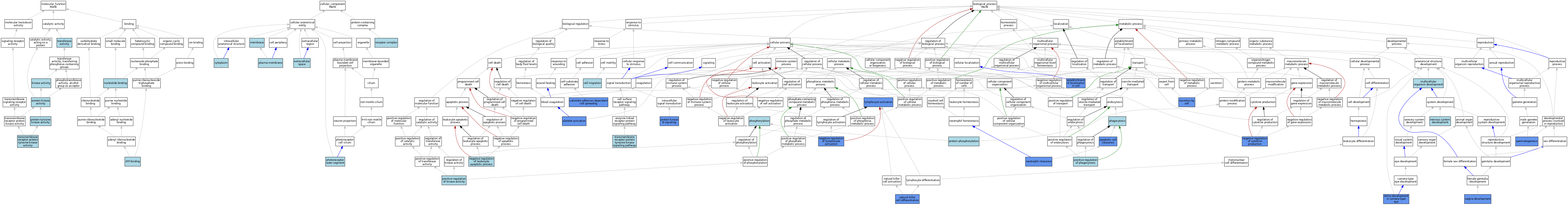
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IMP | J:133455 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IDA | J:265778 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IGI | J:133455 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IGI | J:133455 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IGI | J:200704 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IMP | J:145835 | |||||||||
| Biological Process | GO:0016477 | cell migration | IBA | J:265628 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IMP | J:95952 | |||||||||
| Biological Process | GO:0046649 | lymphocyte activation | IGI | J:70420 | |||||||||
| Biological Process | GO:0046649 | lymphocyte activation | IGI | J:70420 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IBA | J:265628 | |||||||||
| Biological Process | GO:0001779 | natural killer cell differentiation | IGI | J:112666 | |||||||||
| Biological Process | GO:0001779 | natural killer cell differentiation | IGI | J:112666 | |||||||||
| Biological Process | GO:0001818 | negative regulation of cytokine production | IGI | J:200704 | |||||||||
| Biological Process | GO:2000107 | negative regulation of leukocyte apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051250 | negative regulation of lymphocyte activation | IGI | J:70420 | |||||||||
| Biological Process | GO:0051250 | negative regulation of lymphocyte activation | IGI | J:70420 | |||||||||
| Biological Process | GO:0007399 | nervous system development | IBA | J:265628 | |||||||||
| Biological Process | GO:0097350 | neutrophil clearance | IGI | J:200704 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0030168 | platelet activation | IMP | J:95952 | |||||||||
| Biological Process | GO:0033674 | positive regulation of kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0050766 | positive regulation of phagocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0050766 | positive regulation of phagocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0043491 | protein kinase B signaling | IGI | J:95952 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0060041 | retina development in camera-type eye | IMP | J:166679 | |||||||||
| Biological Process | GO:0032940 | secretion by cell | IMP | J:95952 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IGI | J:54681 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IGI | J:54681 | |||||||||
| Biological Process | GO:0034446 | substrate adhesion-dependent cell spreading | IMP | J:95952 | |||||||||
| Biological Process | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0060068 | vagina development | IGI | J:142166 | |||||||||
| Biological Process | GO:0060068 | vagina development | IGI | J:142166 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||