|
Symbol Name ID |
Lhx2
LIM homeobox protein 2 MGI:96785 |
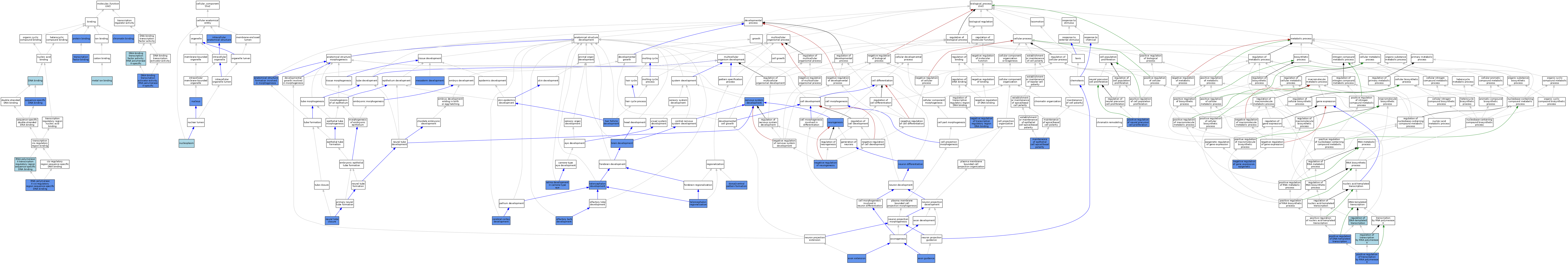
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:145977 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:202029 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | IMP | J:239218 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | IDA | J:239218 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | IMP | J:227000 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:172915 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:237908 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:239195 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:177242 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:49807 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:56497 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IMP | J:237908 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IMP | J:239218 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IDA | J:90847 | |||||||||
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IDA | J:90847 | |||||||||
| Molecular Function | GO:0008134 | transcription factor binding | IPI | J:129101 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | IDA | J:57340 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9011075 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9011082 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9011146 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9753028 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IMP | J:237908 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IMP | J:227000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:130846 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:202029 | |||||||||
| Biological Process | GO:0048646 | anatomical structure formation involved in morphogenesis | IMP | J:73082 | |||||||||
| Biological Process | GO:0048675 | axon extension | IDA | J:179784 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IMP | J:183861 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IMP | J:122567 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:86254 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:91847 | |||||||||
| Biological Process | GO:0021987 | cerebral cortex development | IMP | J:237908 | |||||||||
| Biological Process | GO:0021987 | cerebral cortex development | IMP | J:227000 | |||||||||
| Biological Process | GO:0021987 | cerebral cortex development | IMP | J:154611 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IMP | J:67304 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IMP | J:86254 | |||||||||
| Biological Process | GO:0001942 | hair follicle development | IMP | J:202029 | |||||||||
| Biological Process | GO:0045199 | maintenance of epithelial cell apical/basal polarity | IMP | J:202029 | |||||||||
| Biological Process | GO:0007498 | mesoderm development | IMP | J:64676 | |||||||||
| Biological Process | GO:0045814 | negative regulation of gene expression, epigenetic | IMP | J:237908 | |||||||||
| Biological Process | GO:0050768 | negative regulation of neurogenesis | IMP | J:227000 | |||||||||
| Biological Process | GO:0050768 | negative regulation of neurogenesis | IMP | J:237908 | |||||||||
| Biological Process | GO:2000678 | negative regulation of transcription regulatory region DNA binding | IGI | J:49807 | |||||||||
| Biological Process | GO:0007399 | nervous system development | IMP | J:64676 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IMP | J:73082 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | IDA | J:133461 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:90847 | |||||||||
| Biological Process | GO:0021772 | olfactory bulb development | IMP | J:154611 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:172915 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IMP | J:219181 | |||||||||
| Biological Process | GO:2000179 | positive regulation of neural precursor cell proliferation | IMP | J:227000 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:227000 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:239218 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:145977 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:145977 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:202029 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:56497 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0060041 | retina development in camera-type eye | IGI | J:145977 | |||||||||
| Biological Process | GO:0021537 | telencephalon development | IMP | J:67304 | |||||||||
| Biological Process | GO:0021537 | telencephalon development | IMP | J:122567 | |||||||||
| Biological Process | GO:0021537 | telencephalon development | IMP | J:154611 | |||||||||
| Biological Process | GO:0021537 | telencephalon development | IMP | J:73082 | |||||||||
| Biological Process | GO:0021978 | telencephalon regionalization | IGI | J:73082 | |||||||||
| Biological Process | GO:0021978 | telencephalon regionalization | IGI | J:73082 | |||||||||
| Biological Process | GO:0021978 | telencephalon regionalization | IMP | J:73082 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||