|
Symbol Name ID |
Itgb4
integrin beta 4 MGI:96613 |
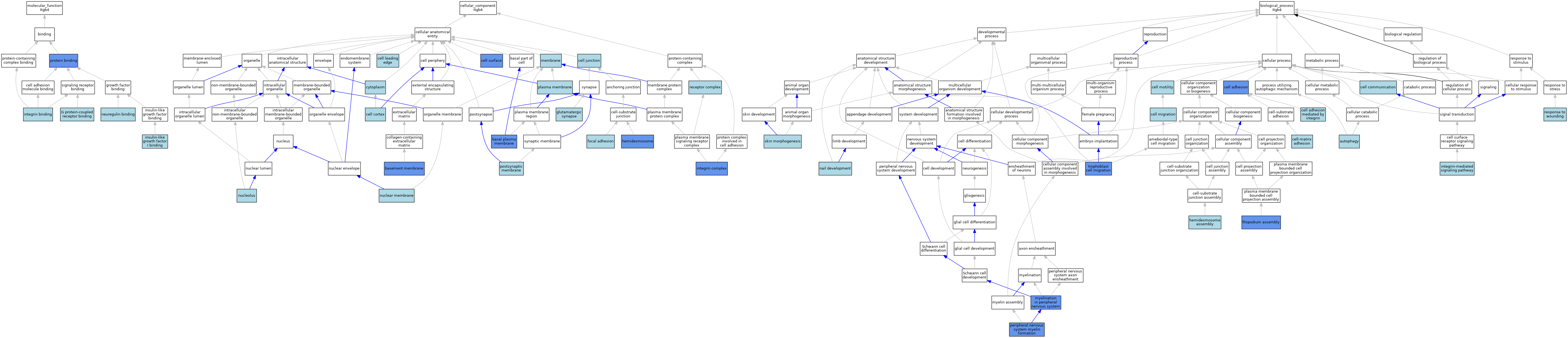
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0001664 | G protein-coupled receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031994 | insulin-like growth factor I binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0038132 | neuregulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:261524 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104989 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:261524 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104989 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104989 | |||||||||
| Cellular Component | GO:0009925 | basal plasma membrane | IDA | J:171374 | |||||||||
| Cellular Component | GO:0009925 | basal plasma membrane | IDA | J:75058 | |||||||||
| Cellular Component | GO:0005604 | basement membrane | IDA | J:116050 | |||||||||
| Cellular Component | GO:0005938 | cell cortex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030054 | cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031252 | cell leading edge | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IBA | J:265628 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:104989 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030056 | hemidesmosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030056 | hemidesmosome | IDA | J:181275 | |||||||||
| Cellular Component | GO:0030056 | hemidesmosome | IDA | J:174237 | |||||||||
| Cellular Component | GO:0008305 | integrin complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0008305 | integrin complex | IDA | J:77644 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | ISO | J:194542 | |||||||||
| Biological Process | GO:0006914 | autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IMP | J:77644 | |||||||||
| Biological Process | GO:0033627 | cell adhesion mediated by integrin | IBA | J:265628 | |||||||||
| Biological Process | GO:0007154 | cell communication | IEA | J:72247 | |||||||||
| Biological Process | GO:0016477 | cell migration | IBA | J:265628 | |||||||||
| Biological Process | GO:0048870 | cell motility | ISO | J:164563 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0046847 | filopodium assembly | IMP | J:108325 | |||||||||
| Biological Process | GO:0031581 | hemidesmosome assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0007229 | integrin-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0022011 | myelination in peripheral nervous system | IGI | J:261524 | |||||||||
| Biological Process | GO:0035878 | nail development | ISO | J:164563 | |||||||||
| Biological Process | GO:0032290 | peripheral nervous system myelin formation | IGI | J:261524 | |||||||||
| Biological Process | GO:0009611 | response to wounding | ISO | J:164563 | |||||||||
| Biological Process | GO:0043589 | skin morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0061450 | trophoblast cell migration | IMP | J:174237 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||