|
Symbol Name ID |
Hbegf
heparin-binding EGF-like growth factor MGI:96070 |
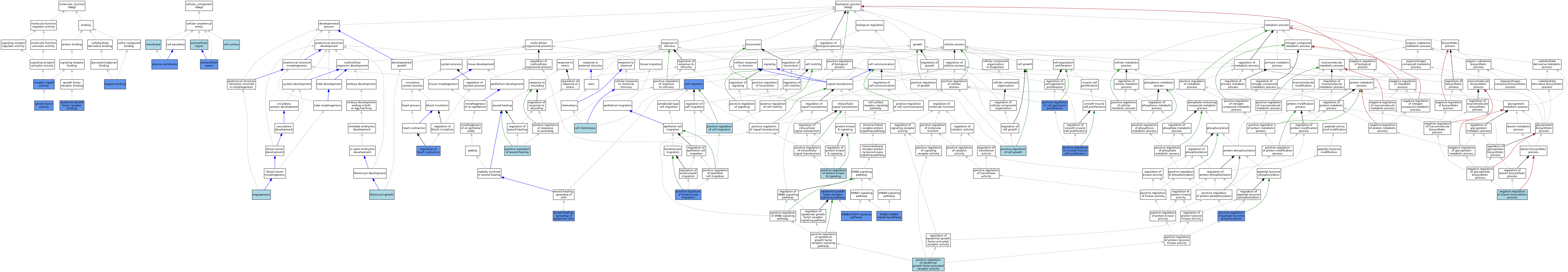
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005154 | epidermal growth factor receptor binding | IDA | J:114809 | |||||||||
| Molecular Function | GO:0005154 | epidermal growth factor receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008083 | growth factor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008083 | growth factor activity | IDA | J:114809 | |||||||||
| Molecular Function | GO:0008083 | growth factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | IDA | J:114809 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IMP | J:177277 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IMP | J:127663 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IMP | J:82728 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IDA | J:82728 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | ISO | J:34905 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-2203911 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:51810 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:177277 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:82728 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:34905 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:51810 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | NAS | J:116267 | |||||||||
| Biological Process | GO:0001832 | blastocyst growth | TAS | J:52356 | |||||||||
| Biological Process | GO:0060326 | cell chemotaxis | ISO | J:164563 | |||||||||
| Biological Process | GO:0016477 | cell migration | IMP | J:99026 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | ISO | J:34905 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IDA | J:82728 | |||||||||
| Biological Process | GO:0038134 | ERBB2-EGFR signaling pathway | IMP | J:177277 | |||||||||
| Biological Process | GO:0038135 | ERBB2-ERBB4 signaling pathway | IMP | J:82728 | |||||||||
| Biological Process | GO:0051545 | negative regulation of elastin biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0030307 | positive regulation of cell growth | ISO | J:155856 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IBA | J:265628 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IDA | J:34905 | |||||||||
| Biological Process | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0051549 | positive regulation of keratinocyte migration | IMP | J:99026 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IDA | J:34905 | |||||||||
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0048661 | positive regulation of smooth muscle cell proliferation | IDA | J:114809 | |||||||||
| Biological Process | GO:0090303 | positive regulation of wound healing | ISO | J:164563 | |||||||||
| Biological Process | GO:0008016 | regulation of heart contraction | IMP | J:82728 | |||||||||
| Biological Process | GO:0035313 | wound healing, spreading of epidermal cells | IMP | J:99026 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||