|
Symbol Name ID |
Glra4
glycine receptor, alpha 4 subunit MGI:95750 |
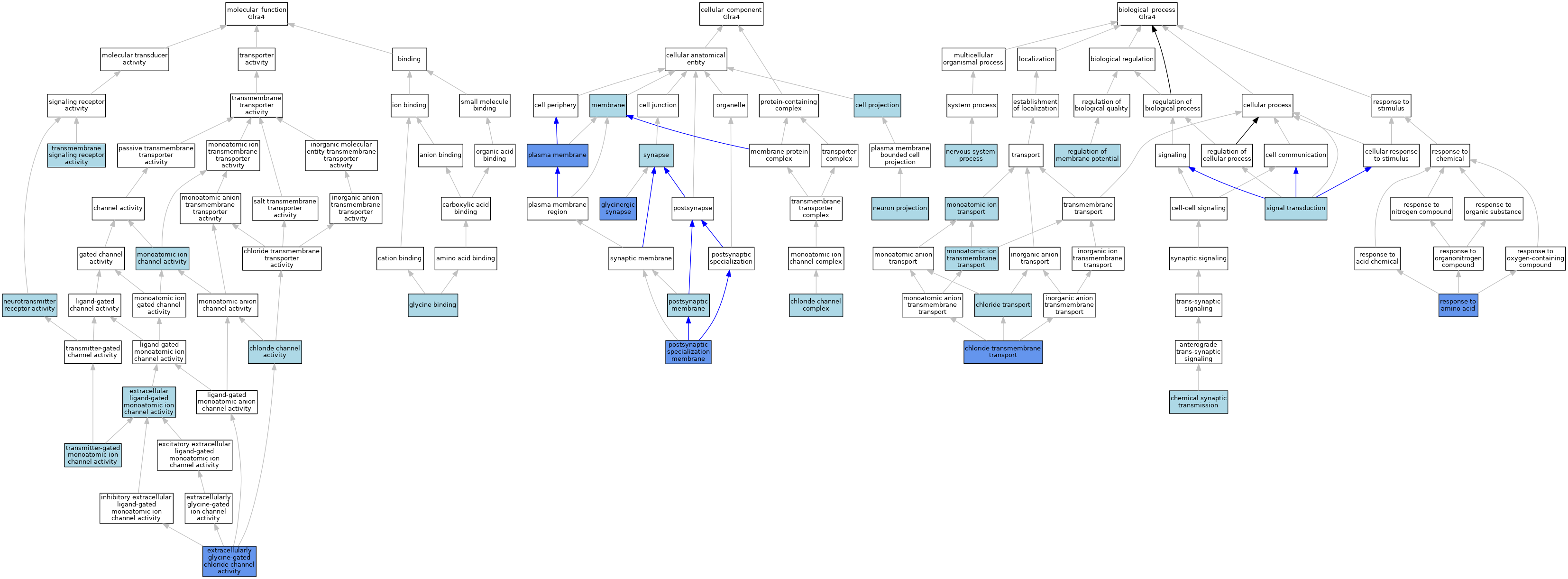
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005254 | chloride channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005230 | extracellular ligand-gated monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016934 | extracellularly glycine-gated chloride channel activity | IDA | J:229376 | |||||||||
| Molecular Function | GO:0016594 | glycine binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005216 | monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0030594 | neurotransmitter receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004888 | transmembrane signaling receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0022824 | transmitter-gated monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0034707 | chloride channel complex | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098690 | glycinergic synapse | IDA | J:263489 | |||||||||
| Cellular Component | GO:0098690 | glycinergic synapse | IDA | J:263489 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:229376 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0099634 | postsynaptic specialization membrane | IDA | J:263489 | |||||||||
| Cellular Component | GO:0099634 | postsynaptic specialization membrane | IDA | J:263489 | |||||||||
| Cellular Component | GO:0045202 | synapse | IBA | J:265628 | |||||||||
| Biological Process | GO:0007268 | chemical synaptic transmission | IBA | J:265628 | |||||||||
| Biological Process | GO:1902476 | chloride transmembrane transport | IDA | J:229376 | |||||||||
| Biological Process | GO:1902476 | chloride transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006821 | chloride transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0034220 | monoatomic ion transmembrane transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0050877 | nervous system process | IBA | J:265628 | |||||||||
| Biological Process | GO:0042391 | regulation of membrane potential | IBA | J:265628 | |||||||||
| Biological Process | GO:0043200 | response to amino acid | IDA | J:229376 | |||||||||
| Biological Process | GO:0043200 | response to amino acid | IBA | J:265628 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||