|
Symbol Name ID |
Glo1
glyoxalase 1 MGI:95742 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
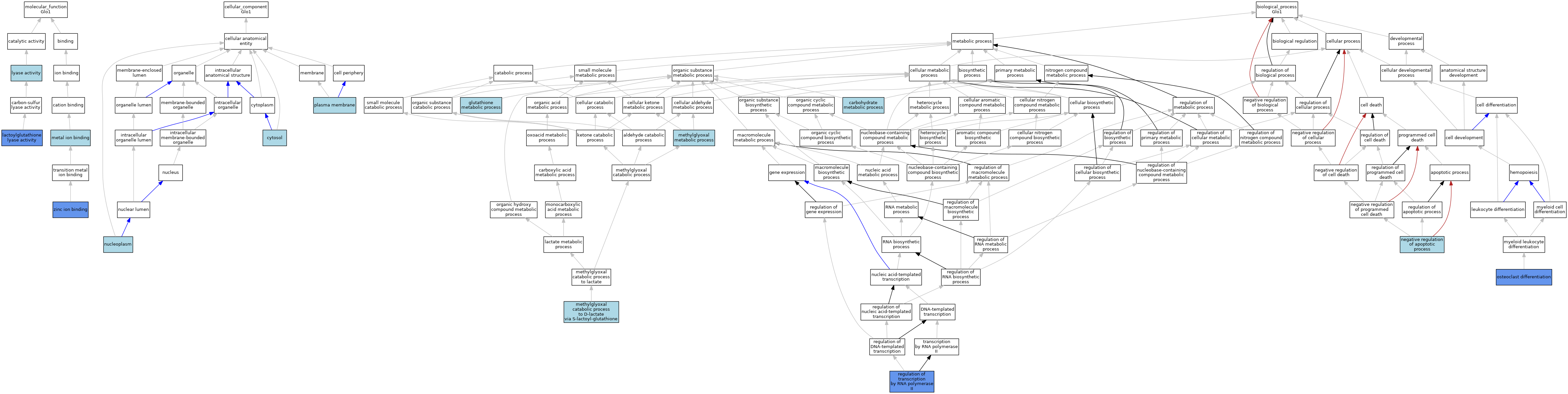
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:203406 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:441 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:178902 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:5998 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:5879 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:6441 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:6704 | |||||||||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | IDA | J:6674 | |||||||||
| Molecular Function | GO:0016829 | lyase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IDA | J:203406 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006749 | glutathione metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | IC | J:178902 | |||||||||
| Biological Process | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | IC | J:6674 | |||||||||
| Biological Process | GO:0009438 | methylglyoxal metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0030316 | osteoclast differentiation | IMP | J:203406 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:126723 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||