|
Symbol Name ID |
Gda
guanine deaminase MGI:95678 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
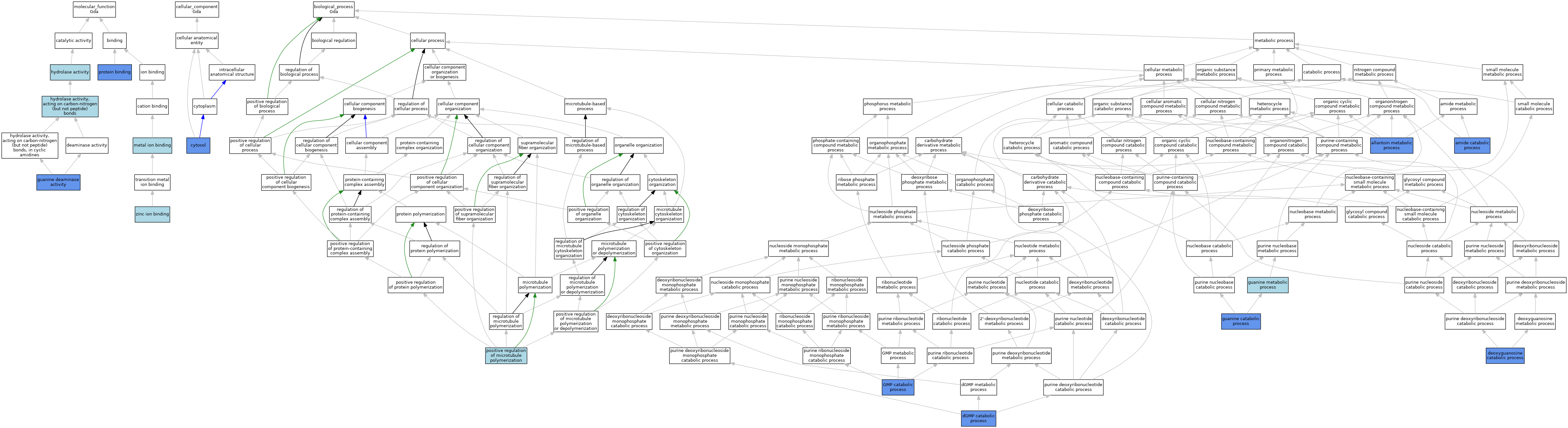
| Molecular Function | GO:0008892 | guanine deaminase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:54278 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297462 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297526 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297526 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297526 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:54278 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297526 | |||||||||
| Molecular Function | GO:0008892 | guanine deaminase activity | IDA | J:297462 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:249721 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:175149 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:152628 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:54278 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:297526 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:54278 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:297526 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:54278 | |||||||||
| Biological Process | GO:0000255 | allantoin metabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0000255 | allantoin metabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0043605 | amide catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0043605 | amide catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0006161 | deoxyguanosine catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0046055 | dGMP catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0046038 | GMP catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0006147 | guanine catabolic process | IDA | J:297526 | |||||||||
| Biological Process | GO:0046098 | guanine metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0031116 | positive regulation of microtubule polymerization | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||