|
Symbol Name ID |
Xrn1
5'-3' exoribonuclease 1 MGI:891964 |
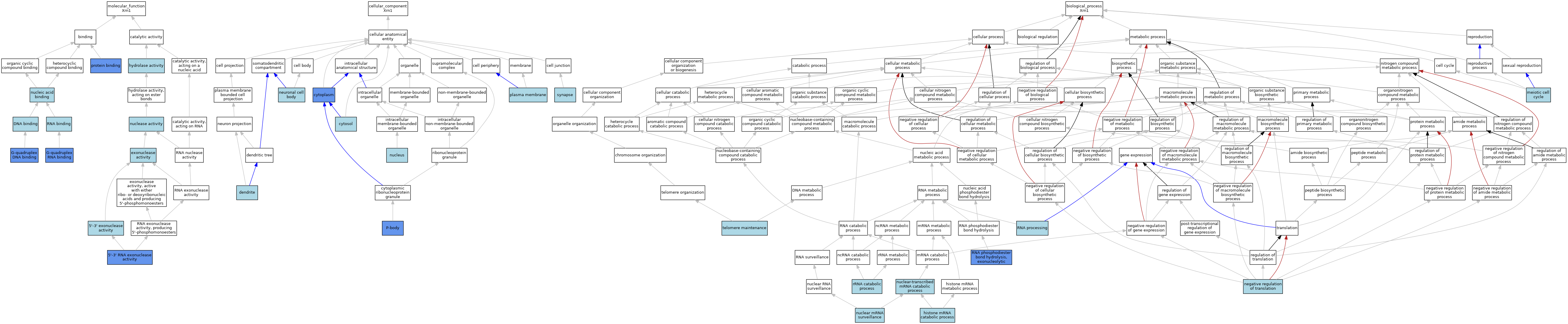
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008409 | 5'-3' exonuclease activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004534 | 5'-3' RNA exonuclease activity | IDA | J:39219 | |||||||||
| Molecular Function | GO:0004534 | 5'-3' RNA exonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004534 | 5'-3' RNA exonuclease activity | TAS | J:41259 | |||||||||
| Molecular Function | GO:0004534 | 5'-3' RNA exonuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004527 | exonuclease activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004527 | exonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051880 | G-quadruplex DNA binding | IDA | J:39219 | |||||||||
| Molecular Function | GO:0002151 | G-quadruplex RNA binding | IDA | J:39219 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:243742 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:39219 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000932 | P-body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0000932 | P-body | IDA | J:139263 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Biological Process | GO:0071044 | histone mRNA catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | NAS | J:41259 | |||||||||
| Biological Process | GO:0017148 | negative regulation of translation | ISO | J:155856 | |||||||||
| Biological Process | GO:0071028 | nuclear mRNA surveillance | ISO | J:164563 | |||||||||
| Biological Process | GO:0000956 | nuclear-transcribed mRNA catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0000956 | nuclear-transcribed mRNA catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | IDA | J:39219 | |||||||||
| Biological Process | GO:0006396 | RNA processing | TAS | J:39219 | |||||||||
| Biological Process | GO:0016075 | rRNA catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0016075 | rRNA catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0000723 | telomere maintenance | NAS | J:41259 | |||||||||
| Biological Process | GO:0000723 | telomere maintenance | IC | J:39219 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||