|
Symbol Name ID |
Chrm3
cholinergic receptor, muscarinic 3, cardiac MGI:88398 |
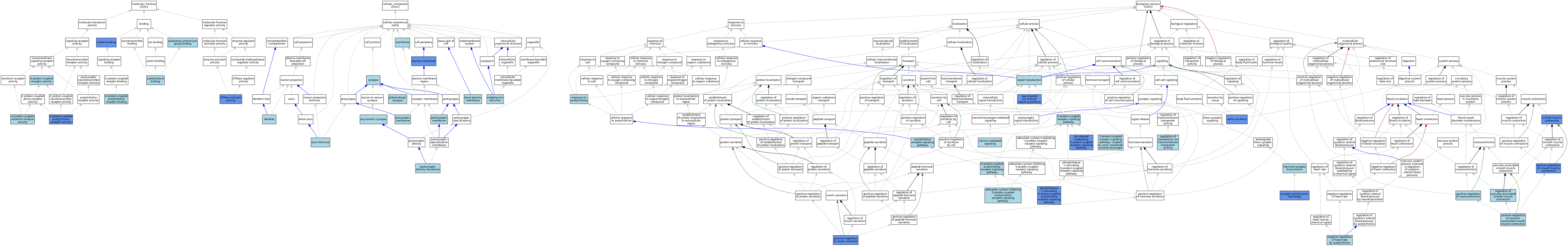
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042166 | acetylcholine binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016907 | G protein-coupled acetylcholine receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016907 | G protein-coupled acetylcholine receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016907 | G protein-coupled acetylcholine receptor activity | IDA | J:161458 | |||||||||
| Molecular Function | GO:0016907 | G protein-coupled acetylcholine receptor activity | IMP | J:129643 | |||||||||
| Molecular Function | GO:0031789 | G protein-coupled acetylcholine receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004993 | G protein-coupled serotonin receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | IDA | J:161458 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | IMP | J:129643 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:233576 | |||||||||
| Molecular Function | GO:0050997 | quaternary ammonium group binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032279 | asymmetric synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043679 | axon terminus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009925 | basal plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-426547 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-426560 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:161458 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IMP | J:129643 | |||||||||
| Cellular Component | GO:0098839 | postsynaptic density membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042734 | presynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IBA | J:265628 | |||||||||
| Biological Process | GO:0095500 | acetylcholine receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007197 | adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0019722 | calcium-mediated signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0007268 | chemical synaptic transmission | IBA | J:265628 | |||||||||
| Biological Process | GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | IBA | J:265628 | |||||||||
| Biological Process | GO:0099105 | ion channel modulating, G protein-coupled receptor signaling pathway | IDA | J:161458 | |||||||||
| Biological Process | GO:1990806 | ligand-gated ion channel signaling pathway | IDA | J:161458 | |||||||||
| Biological Process | GO:0003063 | negative regulation of heart rate by acetylcholine | ISO | J:155856 | |||||||||
| Biological Process | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | IDA | J:161458 | |||||||||
| Biological Process | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | IMP | J:129643 | |||||||||
| Biological Process | GO:0032024 | positive regulation of insulin secretion | IDA | J:161458 | |||||||||
| Biological Process | GO:0032024 | positive regulation of insulin secretion | IMP | J:129643 | |||||||||
| Biological Process | GO:0045987 | positive regulation of smooth muscle contraction | IDA | J:85126 | |||||||||
| Biological Process | GO:1904695 | positive regulation of vascular associated smooth muscle contraction | ISO | J:155856 | |||||||||
| Biological Process | GO:0045907 | positive regulation of vasoconstriction | ISO | J:155856 | |||||||||
| Biological Process | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0003056 | regulation of vascular associated smooth muscle contraction | IBA | J:265628 | |||||||||
| Biological Process | GO:1905144 | response to acetylcholine | ISO | J:155856 | |||||||||
| Biological Process | GO:0046541 | saliva secretion | IMP | J:64069 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0006939 | smooth muscle contraction | IMP | J:64069 | |||||||||
| Biological Process | GO:0006939 | smooth muscle contraction | IDA | J:85126 | |||||||||
| Biological Process | GO:0007271 | synaptic transmission, cholinergic | IMP | J:64069 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||