|
Symbol Name ID |
Ccnd2
cyclin D2 MGI:88314 |
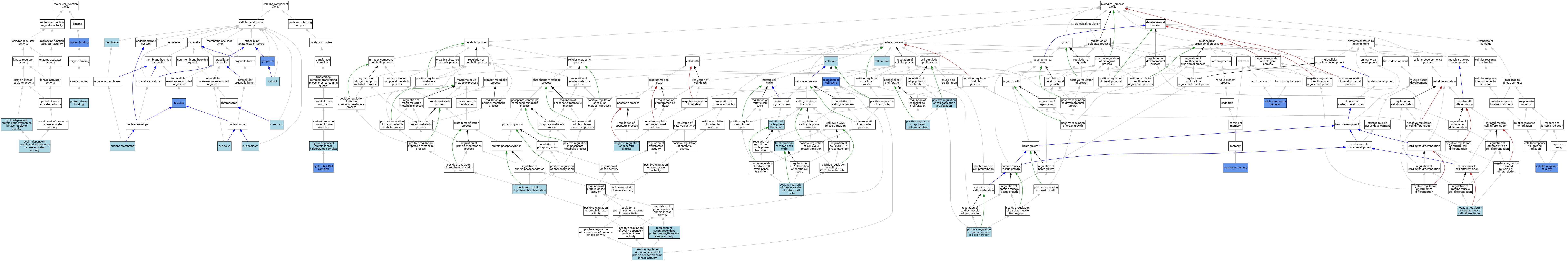
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:19218 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:183053 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097129 | cyclin D2-CDK4 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097129 | cyclin D2-CDK4 complex | IDA | J:24521 | |||||||||
| Cellular Component | GO:0097129 | cyclin D2-CDK4 complex | IDA | J:30542 | |||||||||
| Cellular Component | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:230033 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:85611 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:190888 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:230033 | |||||||||
| Biological Process | GO:0008344 | adult locomotory behavior | IMP | J:199107 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0071481 | cellular response to X-ray | IMP | J:199107 | |||||||||
| Biological Process | GO:0000082 | G1/S transition of mitotic cell cycle | NAS | J:320000 | |||||||||
| Biological Process | GO:0000082 | G1/S transition of mitotic cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0000082 | G1/S transition of mitotic cell cycle | ISO | J:155856 | |||||||||
| Biological Process | GO:0007616 | long-term memory | IMP | J:199107 | |||||||||
| Biological Process | GO:0044772 | mitotic cell cycle phase transition | IBA | J:265628 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:2000726 | negative regulation of cardiac muscle cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0060045 | positive regulation of cardiac muscle cell proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0050679 | positive regulation of epithelial cell proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | IMP | J:74002 | |||||||||
| Biological Process | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||