|
Symbol Name ID |
C1qa
complement component 1, q subcomponent, alpha polypeptide MGI:88223 |
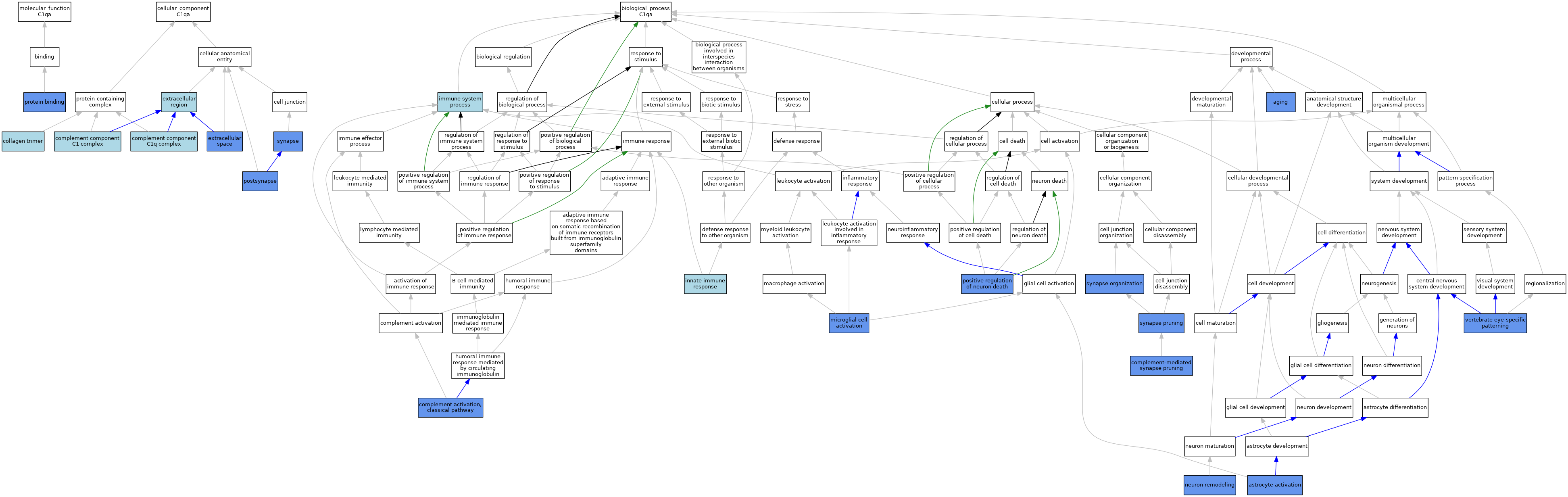
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:186158 | |||||||||
| Cellular Component | GO:0005581 | collagen trimer | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005602 | complement component C1 complex | NAS | J:319954 | |||||||||
| Cellular Component | GO:0062167 | complement component C1q complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | NAS | J:319954 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IGI | J:269645 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:269645 | |||||||||
| Cellular Component | GO:0045202 | synapse | IGI | J:137589 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:137589 | |||||||||
| Biological Process | GO:0007568 | aging | IGI | J:102186 | |||||||||
| Biological Process | GO:0048143 | astrocyte activation | IGI | J:102186 | |||||||||
| Biological Process | GO:0006958 | complement activation, classical pathway | NAS | J:320127 | |||||||||
| Biological Process | GO:0006958 | complement activation, classical pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0006958 | complement activation, classical pathway | IMP | J:75709 | |||||||||
| Biological Process | GO:0150062 | complement-mediated synapse pruning | IMP | J:137589 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0001774 | microglial cell activation | IGI | J:102186 | |||||||||
| Biological Process | GO:0016322 | neuron remodeling | IMP | J:137589 | |||||||||
| Biological Process | GO:1901216 | positive regulation of neuron death | IGI | J:102186 | |||||||||
| Biological Process | GO:0050808 | synapse organization | IGI | J:269645 | |||||||||
| Biological Process | GO:0098883 | synapse pruning | IGI | J:269645 | |||||||||
| Biological Process | GO:0150064 | vertebrate eye-specific patterning | IMP | J:137589 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||