|
Symbol Name ID |
Adss2
adenylosuccinate synthase 2 MGI:87948 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
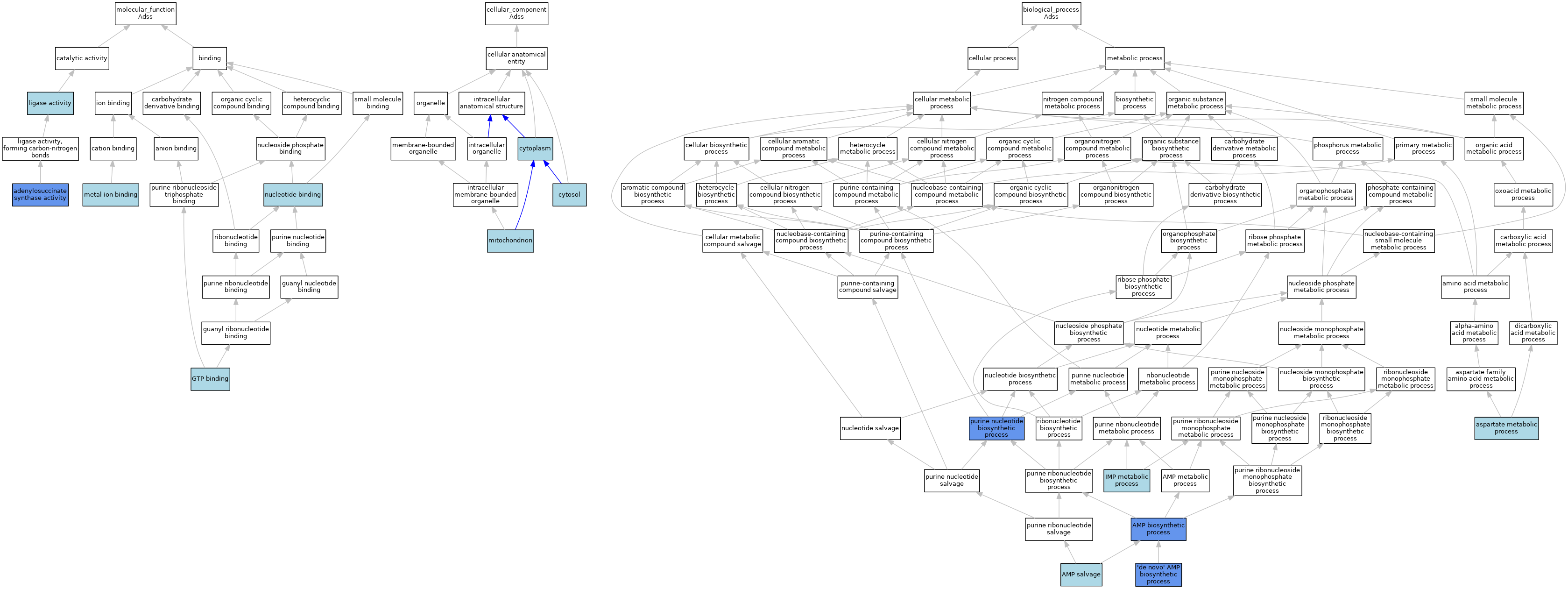
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:11538 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:16814 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:11538 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:82138 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:16814 | |||||||||
| Molecular Function | GO:0004019 | adenylosuccinate synthase activity | IDA | J:82138 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016874 | ligase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-111519 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IEA | J:60000 | |||||||||
| Biological Process | GO:0044208 | 'de novo' AMP biosynthetic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0044208 | 'de novo' AMP biosynthetic process | IMP | J:306719 | |||||||||
| Biological Process | GO:0044208 | 'de novo' AMP biosynthetic process | ISO | J:306656 | |||||||||
| Biological Process | GO:0006167 | AMP biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006167 | AMP biosynthetic process | IDA | J:16814 | |||||||||
| Biological Process | GO:0044209 | AMP salvage | TAS | J:306104 | |||||||||
| Biological Process | GO:0006531 | aspartate metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046040 | IMP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046040 | IMP metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006164 | purine nucleotide biosynthetic process | IDA | J:82138 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||