|
Symbol Name ID |
Sorbs1
sorbin and SH3 domain containing 1 MGI:700014 |
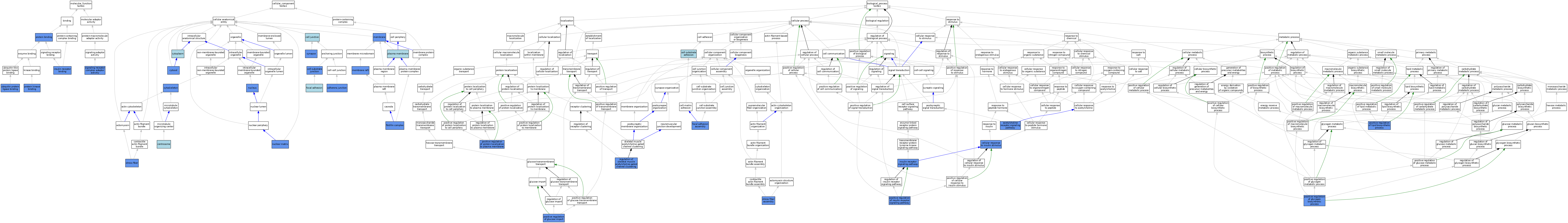
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005158 | insulin receptor binding | IPI | J:46223 | |||||||||
| Molecular Function | GO:0005158 | insulin receptor binding | IDA | J:46223 | |||||||||
| Molecular Function | GO:0005158 | insulin receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114794 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:116379 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:77887 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:45798 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:46223 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200425 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:189995 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:235913 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IDA | J:45798 | |||||||||
| Molecular Function | GO:0030159 | signaling receptor complex adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030159 | signaling receptor complex adaptor activity | IPI | J:46223 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | IPI | J:46223 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IDA | J:116379 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030055 | cell-substrate junction | IDA | J:116379 | |||||||||
| Cellular Component | GO:0030055 | cell-substrate junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IDA | J:189995 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:46223 | |||||||||
| Cellular Component | GO:0016600 | flotillin complex | IDA | J:114794 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:46223 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | IDA | J:114794 | |||||||||
| Cellular Component | GO:0016363 | nuclear matrix | IDA | J:189995 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:189995 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001725 | stress fiber | IDA | J:45798 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:235913 | |||||||||
| Biological Process | GO:0095500 | acetylcholine receptor signaling pathway | IGI | J:235913 | |||||||||
| Biological Process | GO:0095500 | acetylcholine receptor signaling pathway | IGI | J:235913 | |||||||||
| Biological Process | GO:0031589 | cell-substrate adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0032869 | cellular response to insulin stimulus | IMP | J:114794 | |||||||||
| Biological Process | GO:0048041 | focal adhesion assembly | IDA | J:45798 | |||||||||
| Biological Process | GO:0008286 | insulin receptor signaling pathway | IDA | J:46223 | |||||||||
| Biological Process | GO:0046326 | positive regulation of glucose import | IMP | J:114794 | |||||||||
| Biological Process | GO:0046326 | positive regulation of glucose import | IDA | J:114794 | |||||||||
| Biological Process | GO:0045725 | positive regulation of glycogen biosynthetic process | IMP | J:114794 | |||||||||
| Biological Process | GO:0046628 | positive regulation of insulin receptor signaling pathway | IDA | J:114794 | |||||||||
| Biological Process | GO:0046889 | positive regulation of lipid biosynthetic process | IMP | J:114794 | |||||||||
| Biological Process | GO:1903078 | positive regulation of protein localization to plasma membrane | IMP | J:114794 | |||||||||
| Biological Process | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | IGI | J:235913 | |||||||||
| Biological Process | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | IGI | J:235913 | |||||||||
| Biological Process | GO:0043149 | stress fiber assembly | IDA | J:45798 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||